6B6I
 
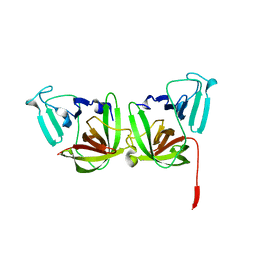 | | 2.4A resolution structure of human Norovirus GII.4 protease | | Descriptor: | 3C-like protease | | Authors: | Muzzarelli, K.M, Kuiper, B.D, Spellmon, N.S, Hackett, J, Brunzelle, J.S, Kovari, I.A, Amblard, F, Yang, Z, Schinazi, R.F, Kovari, L.C. | | Deposit date: | 2017-10-02 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and Antiviral Studies of the Human Norovirus GII.4 Protease.
Biochemistry, 58, 2019
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
7UW2
 
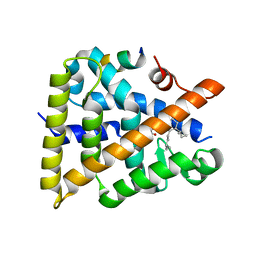 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB116 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-{3-[(2S)-butan-2-yl]-2-(3-methylbutyl)cyclohex-2-en-1-ylidene}-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
7UW4
 
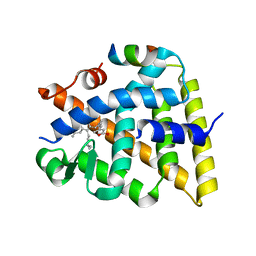 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB113 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-propylcyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
7W82
 
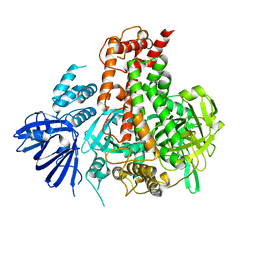 | | Crystal structure of maize RDR2 | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Du, X, Yang, Z, Du, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of plant RNA-DEPENDENT RNA POLYMERASE 2, an enzyme involved in small interfering RNA production.
Plant Cell, 34, 2022
|
|
7W88
 
 | |
7W84
 
 | |
7EER
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with 05E6 and ATP | | Descriptor: | 2-(1H-indol-3-yl)ethanol, ADENOSINE-5'-TRIPHOSPHATE, Tryptophan--tRNA ligase | | Authors: | Lv, G, Fan, S, Feng, X, Zhang, Q, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with O5E6 and ATP
To Be Published
|
|
6BY7
 
 | | Folding DNA into a lipid-conjugated nano-barrel for controlled reconstitution of membrane proteins | | Descriptor: | DNA (26-MER), DNA (27-MER), DNA (29-MER), ... | | Authors: | Dong, Y, Chen, S, Zhang, S, Sodroski, J, Yang, Z, Liu, D, Mao, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7CKI
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lyu, G, Fan, S, Jin, Y, Liu, J, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP
To Be Published
|
|
7CMS
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, PHOSPHATE ION, Tryptophan--tRNA ligase | | Authors: | Lyu, G, Fan, S, Jin, Y, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the specific interaction between Geobacillus stearothermophilus tryptophanyl-tRNA synthetase and antimicrobial Chuangxinmycin.
J.Biol.Chem., 298, 2022
|
|
7CSZ
 
 | | Crystal structure of the N-terminal tandem RRM domains of RBM45 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*GP*GP*AP*CP*GP*C)-3'), RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Wang, W, Wang, M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45.
Nucleic Acids Res., 49, 2021
|
|
7CSX
 
 | |
1XTN
 
 | | crystal structure of CISK-PX domain with sulfates | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase Sgk3 | | Authors: | Xing, Y, Liu, D, Zhang, R, Joachimiak, A, Songyang, Z, Xu, W. | | Deposit date: | 2004-10-22 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX)
J.Biol.Chem., 279, 2004
|
|
1XTE
 
 | | crystal structure of CISK-PX domain | | Descriptor: | Serine/threonine-protein kinase Sgk3 | | Authors: | Xing, Y, Liu, D, Zhang, R, Joachimiak, A, Songyang, Z, Xu, W. | | Deposit date: | 2004-10-21 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX)
J.Biol.Chem., 279, 2004
|
|
8H4R
 
 | | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, G1/S-specific cyclin-E1, ... | | Authors: | Gui, W, Wang, F, Cheng, W, Gao, J, Huang, Y, Ouyang, Z. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus
To Be Published
|
|
5XP0
 
 | |
6KYX
 
 | |
6IJ1
 
 | | Crystal structure of a protein from Actinoplanes | | Descriptor: | ACETATE ION, IMIDAZOLE, Prenylcyclase | | Authors: | Yang, Z.Z, Zhang, L.L, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-10-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Crystal structure of TchmY from Actinoplanes teichomyceticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7RXE
 
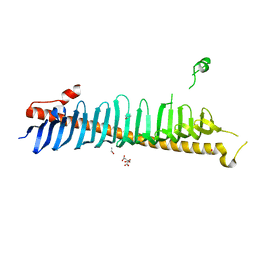 | | Crystal structure of junctophilin-2 | | Descriptor: | CITRATE ANION, ISOPROPYL ALCOHOL, Junctophilin-2 N-terminal fragment | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-22 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RW4
 
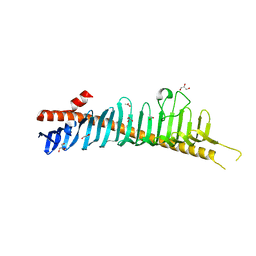 | | Crystal structure of junctophilin-1 | | Descriptor: | ACETATE ION, GLYCEROL, Junctophilin-1 | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RXQ
 
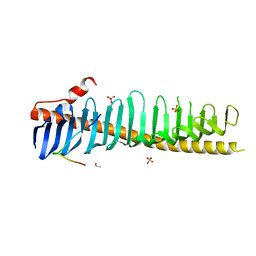 | | Crystal structure of junctophilin-2 in complex with a CaV1.1 peptide | | Descriptor: | ETHANOL, Junctophilin-2 N-terminal fragment, SULFATE ION, ... | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-23 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TLE
 
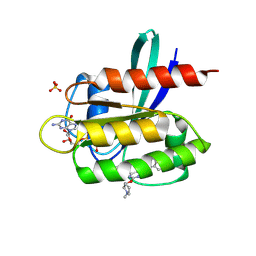 | | Crystal Structure of small molecule beta-lactone 1 covalently bound to K-Ras(G12S) | | Descriptor: | (3R,4R)-1-[7-(8-chloronaphthalen-1-yl)-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]-3-hydroxypiperidine-4-carbaldehyde, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98716748 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
7TLK
 
 | | Crystal Structure of K-Ras(G12S) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71102226 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
7TLG
 
 | | Crystal Structure of small molecule beta-lactone 5 covalently bound to K-Ras(G12S) | | Descriptor: | (3R,4R)-1-[7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(4S,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-4-yl]-3-hydroxypiperidine-4-carbaldehyde, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ziyang, Z, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.80000722 Å) | | Cite: | Chemical acylation of an acquired serine suppresses oncogenic signaling of K-Ras(G12S).
Nat.Chem.Biol., 18, 2022
|
|
