8FAE
 
 | | Asymmetric structure of cleaved HIV-1 AE2 envelope glycoprotein trimer in styrene-maleic acid lipid nanoparticles (AE2.1) | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, K, Zhang, S, Sodroski, J, Mao, Y. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.
Commun Biol, 6, 2023
|
|
8FAD
 
 | | Asymmetric structure of cleaved HIV-1 AD8 envelope glycoprotein trimer in styrene-maleic acid lipid nanoparticles | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, K, Zhang, S, Sodroski, J, Mao, Y. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.
Commun Biol, 6, 2023
|
|
6GN4
 
 | | tc-DNA/tc-DNA duplex | | Descriptor: | Tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3'), Tc-DNA (5'-D(*(TCS)P*(TTK)P*(TCY)P*(TCY)P*(TCS)P*(TCJ)P*(TCJ)P*(TCS)P*(TCY)P*(TCS))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6GMY
 
 | | Tc-DNA/RNA duplex | | Descriptor: | RNA (5'-R(*GP*UP*AP*AP*GP*CP*CP*GP*AP*G)-3'), Tc-DNA (5'-(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6GPI
 
 | | Tc-DNA/DNA duplex | | Descriptor: | DNA (5'-D(*GP*TP*AP*AP*GP*CP*CP*GP*AP*G)-3'), tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-06-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
5JLV
 
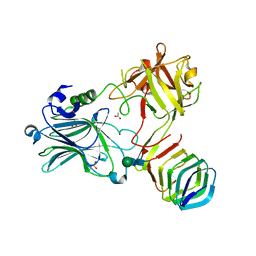 | | Receptor binding domain of Botulinum neurotoxin A in complex with human glycosylated SV2C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6YQX
 
 | | Crystal structure of DeNovoTIM13, a de novo designed TIM barrel | | Descriptor: | CHLORIDE ION, GLYCEROL, de novo designed TIM barrel DeNovoTIM13 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6YQY
 
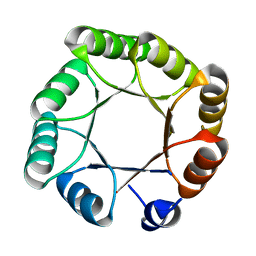 | | Crystal structure of sTIM11noCys, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel sTIM11noCys | | Authors: | Romero-Romero, S, Wiese, G.J, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
3KQ5
 
 | | Crystal structure of an uncharacterized protein from Coxiella burnetii | | Descriptor: | Hypothetical cytosolic protein | | Authors: | Bonanno, J.B, Freeman, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-17 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an uncharacterized protein from Coxiella burnetii
To be Published
|
|
3U9L
 
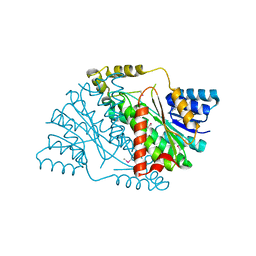 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-19 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
6Z2I
 
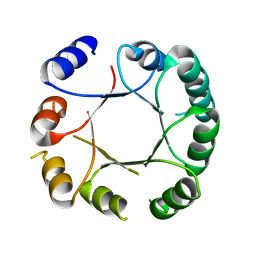 | | Crystal structure of DeNovoTIM6, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel DeNovoTIM6 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-05-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6OYA
 
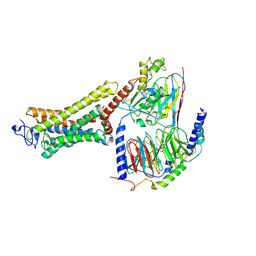 | | Structure of the Rhodopsin-Transducin-Nanobody Complex | | Descriptor: | Camelid antibody VHH fragment, Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
3U9I
 
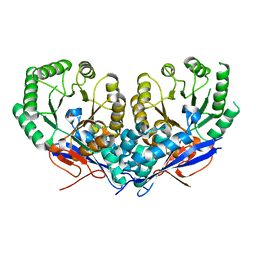 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Roseiflexus sp. | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, C-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-19 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Roseiflexus sp.
TO BE PUBLISHED
|
|
6RLS
 
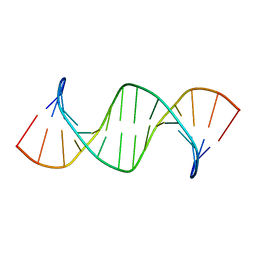 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (apo species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_apo | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
3OMH
 
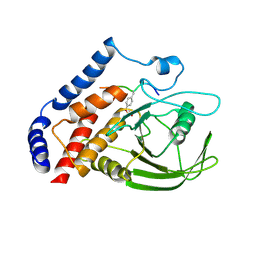 | | Crystal structure of PTPN22 in complex with SKAP-HOM pTyr75 peptide | | Descriptor: | Src kinase-associated phosphoprotein 2, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Yu, X, Sun, J.-P, Zhang, S, Zhang, Z.-Y. | | Deposit date: | 2010-08-26 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate Specificity of Lymphoid-specific Tyrosine Phosphatase (Lyp) and Identification of Src Kinase-associated Protein of 55 kDa Homolog (SKAP-HOM) as a Lyp Substrate.
J.Biol.Chem., 286, 2011
|
|
7T91
 
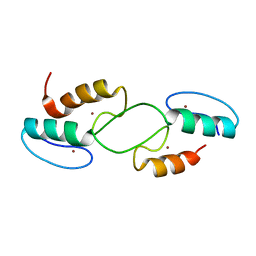 | |
6OY9
 
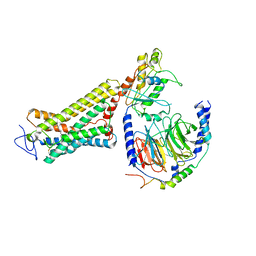 | | Structure of the Rhodopsin-Transducin Complex | | Descriptor: | Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
3SSZ
 
 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium
To be Published
|
|
3T4W
 
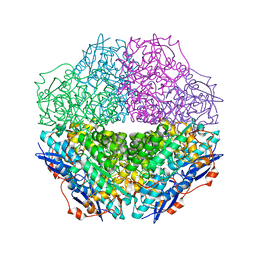 | | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp
To be Published
|
|
7Q48
 
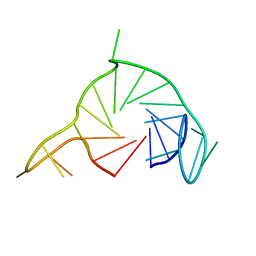 | |
7QA2
 
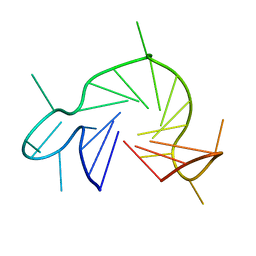 | |
3LQ7
 
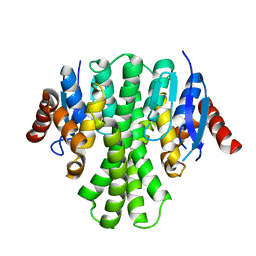 | | Crystal structure of glutathione s-transferase from agrobacterium tumefaciens str. c58 | | Descriptor: | Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Agrobacterium Tumefaciens
To be Published
|
|
5GWM
 
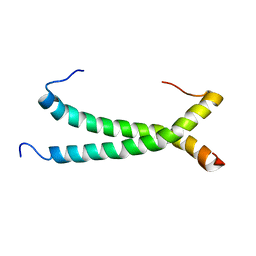 | |
4RIS
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
