5E73
 
 | | Crystal Structure of BAZ2B bromodomain in complex with acetylindole compound UZH47 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)acetamide | | Authors: | Lolli, G, Spiliotopoulos, D, Unzue, A, Nevado, C, Caflisch, A. | | Deposit date: | 2015-10-11 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The "Gatekeeper" Residue Influences the Mode of Binding of Acetyl Indoles to Bromodomains.
J. Med. Chem., 59, 2016
|
|
5E74
 
 | | Crystal Structure of BAZ2B bromodomain in complex with acetylindole compound UZH50 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)-3-(trifluoromethyl)benzamide | | Authors: | Lolli, G, Spiliotopoulos, D, Dolbois, A, Nevado, C, Caflisch, A. | | Deposit date: | 2015-10-11 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The "Gatekeeper" Residue Influences the Mode of Binding of Acetyl Indoles to Bromodomains.
J. Med. Chem., 59, 2016
|
|
6SQM
 
 | | Crystal structure of CREBBP bromodomain complexed with LA36 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-acetamido-5-[(3-methylcinnolin-5-yl)carbamoyl]phenyl]furan-2-carboxamide | | Authors: | Bedi, R.K, Laul, E, Nevado, C, Caflisch, A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexed with LA36
To Be Published
|
|
6SQE
 
 | | Crystal structure of CREBBP bromodomain complexed with KD341 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-[(5-ethanoyl-2-ethoxy-phenyl)carbamoyl]-5-(1-methylpyrazol-3-yl)phenyl]-5-[(4-methylpiperazin-1-yl)methyl]furan-2-carboxamide | | Authors: | Bedi, R.K, Kirillova, M, Nevado, C, Caflisch, A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexed with KD341
To Be Published
|
|
6SQF
 
 | |
6FGL
 
 | | Crystal Structure of BAZ2A bromodomain in complex with acetylindole compound UZH47 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)acetamide | | Authors: | Dalle Vedove, A, Unzue, A, Nevado, C, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
4TQN
 
 | |
5EIC
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with AYC | | Descriptor: | 1,2-ETHANEDIOL, 2-[(chloroacetyl)amino]-5-[(E)-(4-sulfophenyl)diazenyl]benzenesulfonic acid, CREB-binding protein | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5ENG
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with UP39 | | Descriptor: | CREB-binding protein, methyl 2-[2-(3,5-dihydro-2~{H}-pyrazin-4-yl)ethoxy]-5-[(5-ethanoyl-2-ethoxy-phenyl)carbamoyl]benzoate | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5EP7
 
 | |
5F36
 
 | |
5H85
 
 | |
5D7X
 
 | |
4P5Z
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-[4-({[3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl]-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4P5Q
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(2-chlorophenyl)-N-(3-ethoxypropyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4P4C
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(3-methoxyphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4G2F
 
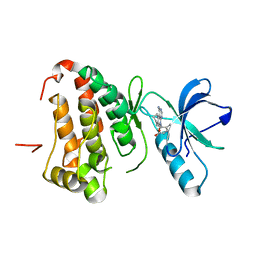 | | Human EphA3 kinase domain in complex with compound 7 | | Descriptor: | 1-amino-5-(5-hydroxy-2-methylphenyl)-7,8,9,10-tetrahydropyrimido[4,5-c]isoquinolin-6(5H)-one, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Discovery of a novel chemotype of tyrosine kinase inhibitors by fragment-based docking and molecular dynamics.
ACS MED.CHEM.LETT., 3, 2012
|
|
4GK3
 
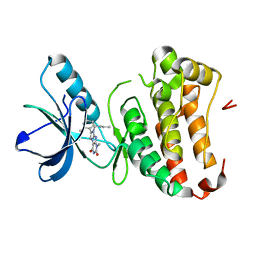 | | Human EphA3 Kinase domain in complex with ligand 87 | | Descriptor: | 8-butyl-1-methyl-7-(2-methylphenyl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
4GK2
 
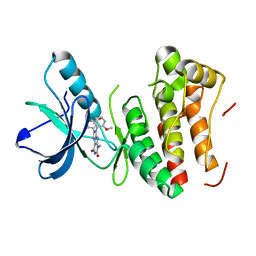 | | Human EphA3 Kinase domain in complex with ligand 66 | | Descriptor: | 7-(5-hydroxy-2-methylphenyl)-8-(2-methoxyphenyl)-1-methyl-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
4GK4
 
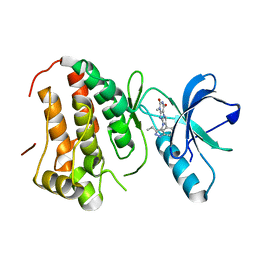 | | Human EphA3 Kinase domain in complex with ligand 90 | | Descriptor: | 8-butyl-1-methyl-7-(5-methyl-1H-indazol-4-yl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
5MME
 
 | | Crystal structure of CREBBP bromodomain complexd with US46C | | Descriptor: | CREB-binding protein, dimethyl 5-[(5-ethanoyl-2-ethoxy-phenyl)amino]benzene-1,3-dicarboxylate | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MPK
 
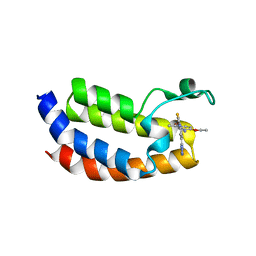 | | Crystal structure of CREBBP bromodomain complexed with DK19 | | Descriptor: | CREB-binding protein, ~{N}-(5-ethanoyl-2-ethoxy-phenyl)-3-(2~{H}-1,2,3,4-tetrazol-5-yl)-5-(1,3-thiazol-4-yl)benzamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MMG
 
 | | Crystal structure of CREBBP bromodomain complexed with UT07C | | Descriptor: | 1-[4-ethoxy-3-[(1-methylsulfonylindol-6-yl)amino]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5MPN
 
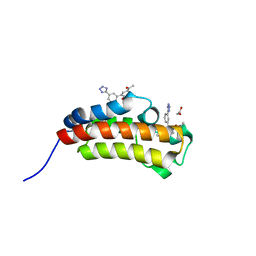 | | Crystal structure of CREBBP bromodomain complexed with FA26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethoxy-3-[3-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5NLK
 
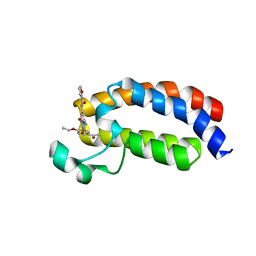 | | Crystal structure of CREBBP bromodomain complexd with US13A | | Descriptor: | CREB-binding protein, ~{N}-[3-acetamido-5-[(5-ethanoyl-2-ethoxy-phenyl)carbamoyl]phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-04-04 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
ACS Cent Sci, 4, 2018
|
|
