5KSJ
 
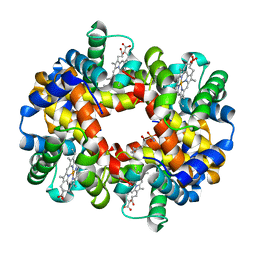 | | Crystal structure of deoxygenated hemoglobin in complex with Sphingosine phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
5NDF
 
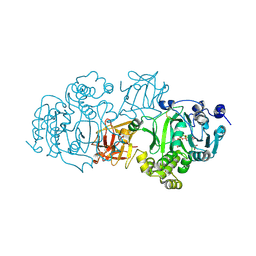 | | Small-molecule inhibition of ppGalNAc-Ts selectively reduces mucin-type O-glycosylation | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, MANGANESE (II) ION, ... | | Authors: | Hurtado-Guerrero, R, De las Rivas, M. | | Deposit date: | 2017-03-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The small molecule luteolin inhibits N-acetyl-alpha-galactosaminyltransferases and reduces mucin-type O-glycosylation of amyloid precursor protein.
J. Biol. Chem., 292, 2017
|
|
6VYC
 
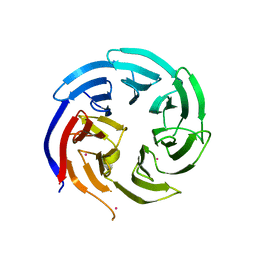 | | Crystal structure of WD-repeat domain of human WDR91 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Halabelian, L, Hutchinson, A, Li, Y, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
6W5I
 
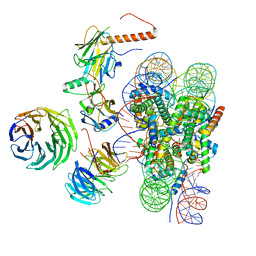 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5N
 
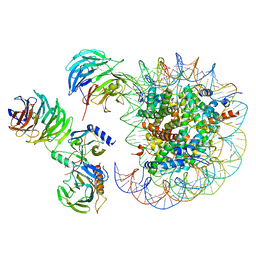 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5M
 
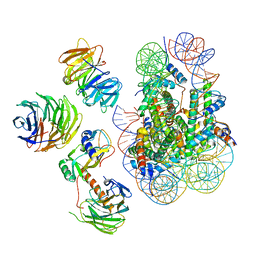 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6QSW
 
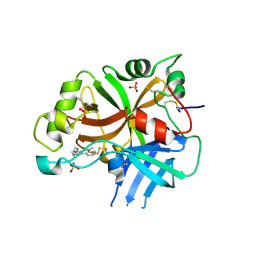 | | Complement factor B protease domain in complex with the reversible inhibitor N-(2-bromo-4-methylnaphthalen-1-yl)-4,5-dihydro-1H-imidazol-2-amine. | | Descriptor: | Complement factor B, SULFATE ION, ~{N}-(2-bromanyl-4-methyl-naphthalen-1-yl)-4,5-dihydro-1~{H}-imidazol-2-amine | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QSX
 
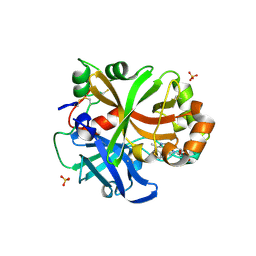 | | Complement factor B protease domain in complex with the reversible inhibitor ((2S,4S)-1-((5,7-dimethyl-1H-indol-4-yl)methyl)-4-methoxypiperidin-2-yl)methanol. | | Descriptor: | Complement factor B, SULFATE ION, ZINC ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2I47
 
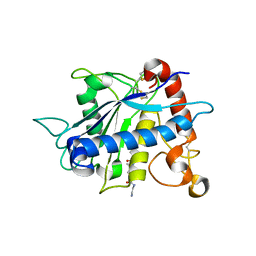 | | Crystal structure of catalytic domain of TACE with inhibitor | | Descriptor: | 4-({[4-(BUT-2-YN-1-YLOXY)PHENYL]SULFONYL}METHYL)-1-[(3,5-DIMETHYLISOXAZOL-4-YL)SULFONYL]-N-HYDROXYPIPERIDINE-4-CARBOXAMIDE, ADAM 17, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Xu, W, Condon, J.S, Lovering, F.E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-12-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of potent and selective TACE inhibitors via the S1 pocket.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6P9Y
 
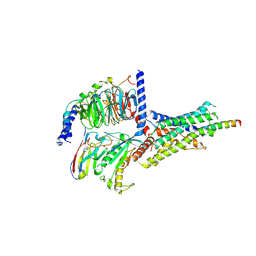 | | PAC1 GPCR Receptor complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-06-10 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation.
Mol.Cell, 77, 2020
|
|
7O5N
 
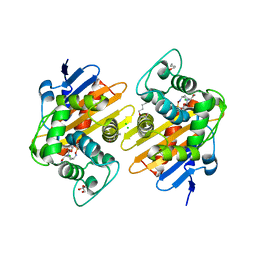 | | Crystal Structure of a Class D carbapenemase complexed with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1-BUTANOL, Beta-lactamase, ... | | Authors: | Zhou, Q, Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
6PNX
 
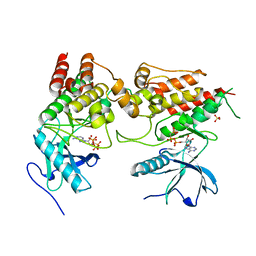 | |
6W45
 
 | | Crystal structure of HAO1 in complex with biaryl acid inhibitor - compound 3 | | Descriptor: | 2-chloranyl-4-[2-[[(6-chloranyl-1~{H}-indol-2-yl)carbonyl-methyl-amino]methyl]-5-fluoranyl-phenyl]benzoic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W44
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 4 | | Descriptor: | 5-[methyl-[(2-propoxypyridin-3-yl)methyl]amino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W4C
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 5 | | Descriptor: | 5-[[3-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-2-oxidanyl-phenyl]methylamino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6PXG
 
 | |
5JYY
 
 | | Structure-based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-resistant Influenza Viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-[(2-methoxyethyl)carbamoyl]-D-glycero-D-galacto-non-2-enon ic acid, CALCIUM ION, ... | | Authors: | Fu, L, Wu, Y, Bi, Y, Zhang, S, Lv, X, Qi, J, Li, Y, Lu, X, Yan, J, Gao, G.F, Li, X. | | Deposit date: | 2016-05-15 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-Resistant Influenza Viruses
J.Med.Chem., 59, 2016
|
|
6ZOV
 
 | | ENTEROPEPTIDASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-carbamimidamidobenzoic acid, Enteropeptidase, ... | | Authors: | Cummings, M.D. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeting Enteropeptidase with Reversible Covalent Inhibitors To Achieve Metabolic Benefits.
J.Pharmacol.Exp.Ther., 375, 2020
|
|
4Y5O
 
 | | CCM2 HHD in complex with MEKK3 NPB1 | | Descriptor: | Malcavernin, Mitogen-activated protein kinase kinase kinase 3 | | Authors: | Fisher, O.S, Boggon, T.J. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex.
Nat Commun, 6, 2015
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
3X05
 
 | | Crystal structure of PIP4KIIBETA T201M complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X06
 
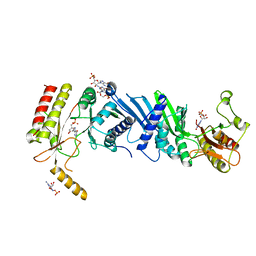 | | Crystal structure of PIP4KIIBETA T201M complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X03
 
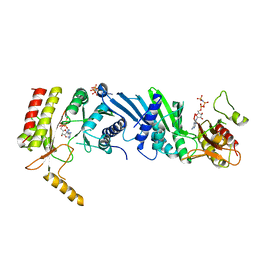 | | Crystal structure of PIP4KIIBETA complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X01
 
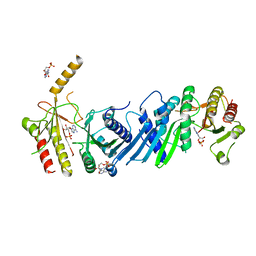 | | Crystal structure of PIP4KIIBETA complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
