7LFI
 
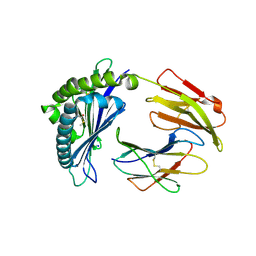 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE REFINED AT 1.70 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFJ
 
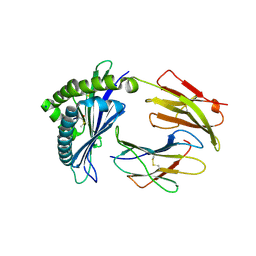 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, ALA MUTANT, REFINED AT 1.70 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
8J6J
 
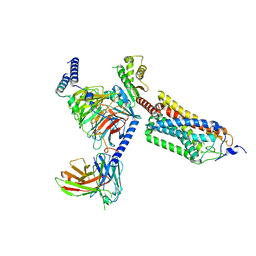 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound with GSK256073 | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8J6L
 
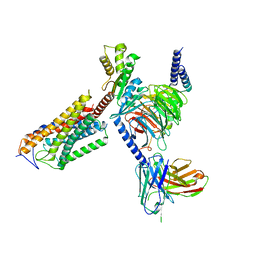 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
6MR5
 
 | |
8J6I
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8GRD
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, ... | | Authors: | Chen, X, Sun, P, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GS5
 
 | | Crystal structure of a constitutively active mutant of human IDH3 holoenzyme in apo form | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.486 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRG
 
 | |
8GRH
 
 | | Crystal structure of a constitutively active mutant of the alpha gamma heterodimer of human IDH3 in complex with CIT | | Descriptor: | CITRIC ACID, Human IDH3 alpha subunit, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRU
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ICT, NAD and Ca | | Descriptor: | CALCIUM ION, Human IDH3 alpha subunit, ISOCITRIC ACID, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRB
 
 | |
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AAS
 
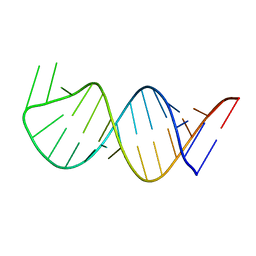 | | Solution Structure for helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (28-MER) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
7WP1
 
 | | Cryo-EM structure of SARS-CoV-2 Mu S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WON
 
 | | Cryo-EM structure of SARS-CoV-2 S2P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP2
 
 | | Cryo-EM structure of SARS-CoV-2 C.1.2 S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP0
 
 | | Cryo-EM structure of SARS-CoV-2 Delta S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c335_light_IGLV1-40_IGLJ3, Spike protein S1, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
8WIL
 
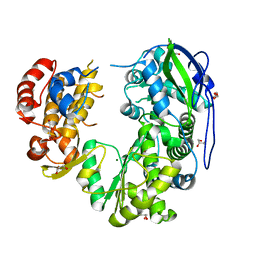 | | Crystal structure of Jingmen tick virus RNA-dependent RNA polymerase (D55 construct) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Jingmen tick virus NSP1, ... | | Authors: | Wang, X, Jing, X, Deng, F, Gong, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A jingmenvirus RNA-dependent RNA polymerase structurally resembles the flavivirus counterpart but with different features at the initiation phase.
Nucleic Acids Res., 52, 2024
|
|
8WIM
 
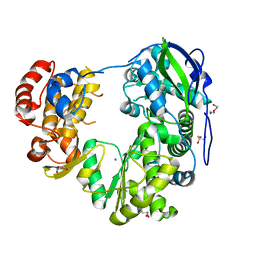 | | Crystal structure of Jingmen tick virus RNA-dependent RNA polymerase (D307 construct) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Jingmen tick virus NSP1, ... | | Authors: | Wang, X, Jing, X, Deng, F, Gong, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A jingmenvirus RNA-dependent RNA polymerase structurally resembles the flavivirus counterpart but with different features at the initiation phase.
Nucleic Acids Res., 52, 2024
|
|
8X3R
 
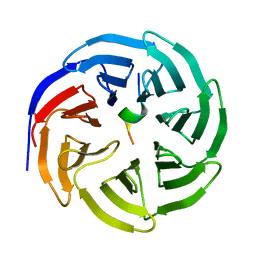 | | Crystal structure of human WDR5 in complex with WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
8X3S
 
 | | Crystal structure of human WDR5 in complex with PTEN | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X, Shang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
8J5Z
 
 | | The cryo-EM structure of the TwOSC1 tetramer | | Descriptor: | Terpene cyclase/mutase family member, octyl beta-D-glucopyranoside | | Authors: | Ma, X, Yuru, T, Yunfeng, L, Jiang, T. | | Deposit date: | 2023-04-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7EVR
 
 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
