7LK1
 
 | | Ornithine Aminotransferase (OAT) with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) - 1 Hour Soaking | | Descriptor: | (1R,4R)-4-fluoro-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-2-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LK0
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) | | Descriptor: | (1R,3S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
6V8D
 
 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | Descriptor: | (3Z)-3-iminocyclohex-1-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | Deposit date: | 2019-12-10 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
6V8C
 
 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | Descriptor: | 3-aminocyclohexa-1,3-diene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | Deposit date: | 2019-12-10 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
8EZ1
 
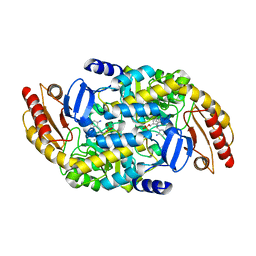 | | Human Ornithine Aminotransferase (hOAT) co-crystallized with its inactivator 3-Amino-4-fluorocyclopentenecarboxylic Acid | | Descriptor: | (1R,3S,4Z)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, (3E,4E)-4-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-3-iminocyclopent-1-ene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Shen, S, Silverman, R, Liu, D. | | Deposit date: | 2022-10-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Mechanistic Basis for the Inactivation of Human Ornithine Aminotransferase by (3 S ,4 S )-3-Amino-4-fluorocyclopentenecarboxylic Acid.
Molecules, 28, 2023
|
|
6AAX
 
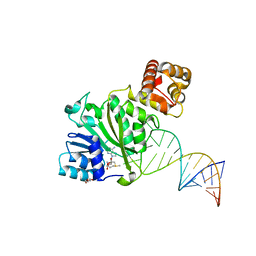 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AJK
 
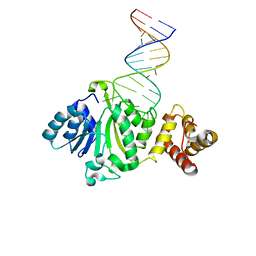 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
7CNC
 
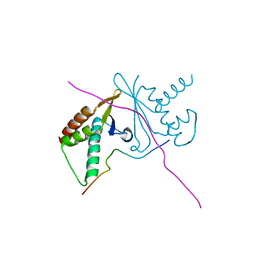 | | cystal structure of human ERH in complex with DGCR8 | | Descriptor: | Enhancer of rudimentary homolog, Microprocessor complex subunit DGCR8 | | Authors: | Li, F, Shen, S. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8.
Nucleic Acids Res., 48, 2020
|
|
7JX9
 
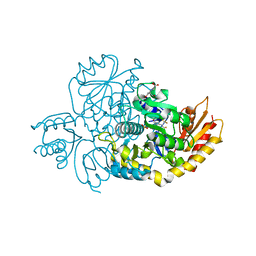 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
7LFM
 
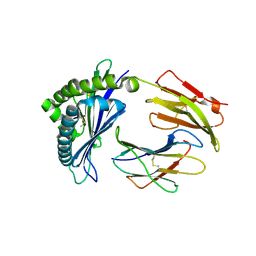 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, TRICLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFJ
 
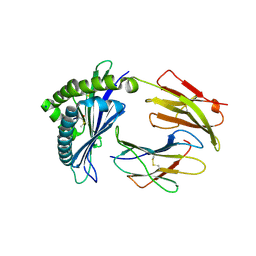 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, ALA MUTANT, REFINED AT 1.70 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFL
 
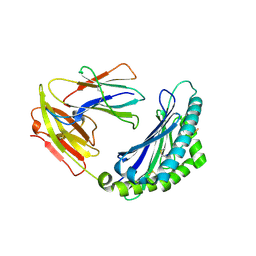 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, MONOCLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFI
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE REFINED AT 1.70 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFK
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, THR MUTANT, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
