6IO4
 
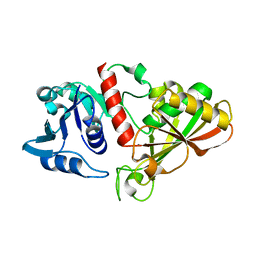 | |
5Z1R
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
2MVT
 
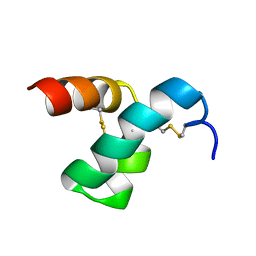 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | Descriptor: | Scoloptoxin SSD609 | | Authors: | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
2MT4
 
 | |
7T7A
 
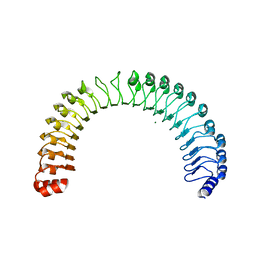 | | Crystal Structure of Human SHOC2: A Leucine-Rich Repeat Protein | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, NITRATE ION | | Authors: | Hajian, B, Lemke, C, Kwon, J, Bian, Y, Fuller, C, Aguirre, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex.
Nature, 609, 2022
|
|
7UPI
 
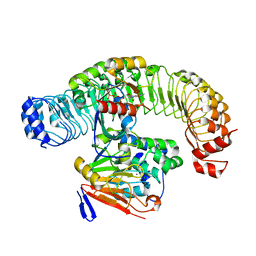 | | Cryo-EM structure of SHOC2-PP1c-MRAS holophosphatase complex | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, Leucine-rich repeat protein SHOC-2, ... | | Authors: | Fuller, J.R, Hajian, B, Lemke, C, Kwon, J, Bian, Y, Aguirre, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex.
Nature, 609, 2022
|
|
8WKJ
 
 | |
8WOU
 
 | |
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
8JWO
 
 | |
8JWN
 
 | | Crystal structure of AKRtyl-NADPH complex | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWL
 
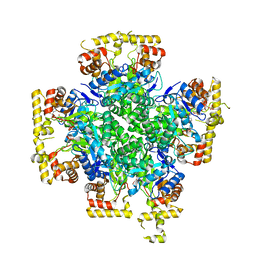 | |
8JWK
 
 | | The second purified state crystal structure of AKRtyl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWM
 
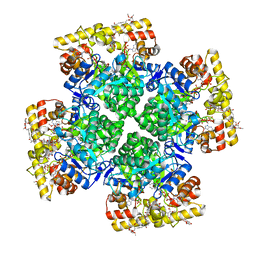 | | Crystal structure of AKRtyl-NADP-tylosin complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TYLOSIN | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8IYJ
 
 | | Cryo-EM structure of the 48-nm repeat doublet microtubule from mouse sperm | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhou, L.N, Gui, M, Wu, J.P. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of sperm flagellar doublet microtubules expand the genetic spectrum of male infertility.
Cell, 186, 2023
|
|
6DD6
 
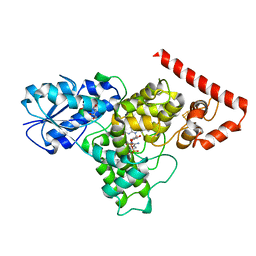 | |
5VK3
 
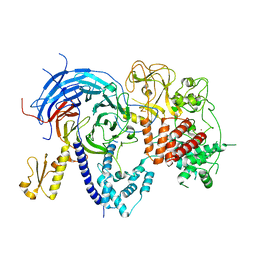 | | Apo ctPRC2 with E840A and K852D mutations in Ezh2 | | Descriptor: | Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12, Polycomb Protein EED, ZINC ION | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-04-20 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
7YVR
 
 | | Crystal Structure of L-Threonine Aldolase from Neptunomonas marina | | Descriptor: | GLYCEROL, L-threonine aldolase | | Authors: | He, Y.Z, Wang, J, Yan, W.P, Zhang, Y, Feng, Y. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Engineering of the L-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of beta-Hydroxy-alpha-amino Acids via C-C Formation
Acs Catalysis, 2023
|
|
5WAK
 
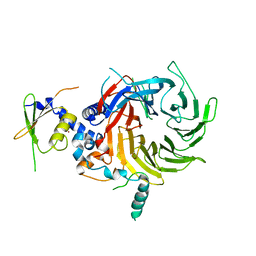 | | Crystal Structure of a Suz12-Rbbp4 Binary Complex | | Descriptor: | Histone-binding protein RBBP4, Polycomb protein SUZ12, ZINC ION | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Mol. Cell, 69, 2018
|
|
5WAI
 
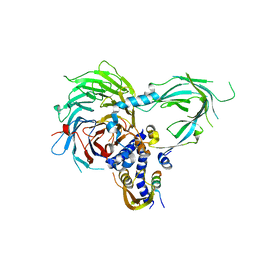 | | Crystal Structure of a Suz12-Rbbp4-Jarid2-Aebp2 Heterotetrameric Complex | | Descriptor: | Histone-binding protein RBBP4, Jumonji, AT-rich interactive domain 2, ... | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Mol. Cell, 69, 2018
|
|
5WFC
 
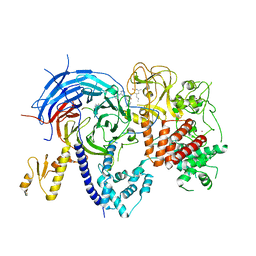 | |
5WFD
 
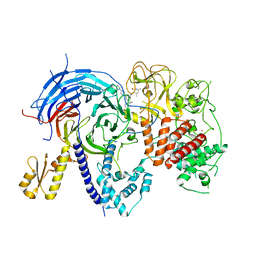 | | Humanized mutant of the Chaetomium thermophilum Polycomb Repressive Complex 2 bound to the inhibitor GSK126 | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine-N-methyltransferase EZH2, Polycomb protein SUZ12 chimera, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
5WF7
 
 | | Chaetomium thermophilum Polycomb Repressive Complex 2 bound to GSK126 | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine-N-methyltransferase EZH2, Polycomb protein SUZ12 chimera, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
6DD7
 
 | |
5WG6
 
 | | Human Polycomb Repressive Complex 2 in complex with GSK126 inhibitor | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12 (E.C.2.1.1.43) chimera, Polycomb protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
