4RFE
 
 | | Crystal structure of ADCC-potent ANTI-HIV-1 Rhesus macaque antibody JR4 Fab | | Descriptor: | CHLORIDE ION, Fab heavy chain of ADCC-potent anti-HIV-1 antibody JR4, Fab light chain of ADCC-potent anti-HIV-1 antibody JR4, ... | | Authors: | Wu, X, Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4RFO
 
 | | Crystal structure of the ADCC-Potent Antibody N60-I3 Fab in complex with HIV-1 Clade A/E gp120 and M48u1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade A/E gp120, N60-i3 Fab heavy chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4RFN
 
 | | Crystal structure of ADCC-potent Rhesus macaque ANTIBODY JR4 in complex with HIV-1 CLADE A/E GP120 and M48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, FAB LIGHT CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
6KEY
 
 | |
7S3I
 
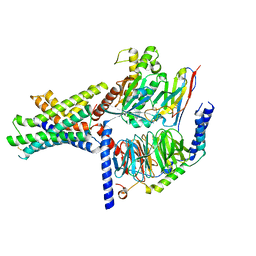 | | Ex4-D-Ala bound to the glucagon-like peptide-1 receptor/g protein complex (conformer 2) | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Piper, S.J, Danev, R. | | Deposit date: | 2021-09-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural and functional diversity among agonist-bound states of the GLP-1 receptor.
Nat.Chem.Biol., 18, 2022
|
|
5XGM
 
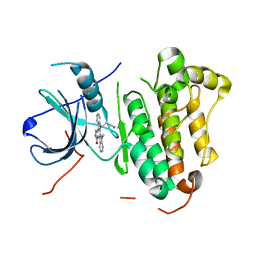 | | Crystal structure of EGFR 696-1022 T790M in complex with Go6976 | | Descriptor: | 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, Epidermal growth factor receptor | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2017-04-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural pharmacological studies on EGFR T790M/C797S.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
5XDK
 
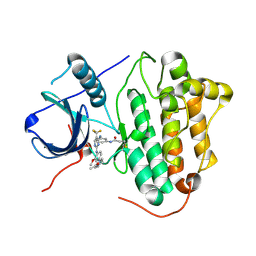 | | Crystal structure of EGFR 696-1022 T790M in complex with CO-1686 | | Descriptor: | Epidermal growth factor receptor, N-[3-[[2-[[4-(4-ethanoylpiperazin-1-yl)-2-methoxy-phenyl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2017-03-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | Structural basis of mutant-selectivity and drug-resistance related to CO-1686.
Oncotarget, 8, 2017
|
|
5XGN
 
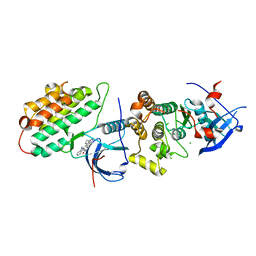 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with Go6976 | | Descriptor: | 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2017-04-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural pharmacological studies on EGFR T790M/C797S.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
4QN0
 
 | | Crystal structure of the CPS-6 mutant Q130K | | Descriptor: | Endonuclease G, mitochondrial, MAGNESIUM ION | | Authors: | Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Oxidative Stress Impairs Cell Death by Repressing the Nuclease Activity of Mitochondrial Endonuclease G
Cell Rep, 16, 2016
|
|
3UP1
 
 | | Crystal structure of the unliganded human interleukin-7 receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | McElroy, C.A, Holland, P.J, Walsh, S.T.R. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural reorganization of the interleukin-7 signaling complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6JXT
 
 | |
5V7J
 
 | | Crystal Structure at 3.7 A Resolution of Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer with Four Glycans (N197, N276, N362, and N462) removed in Complex with Neutralizing Antibodies 3H+109L and 35O22. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 35O22 Fab heavy chain, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Kwong, P.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation.
Cell Rep, 19, 2017
|
|
5X2A
 
 | |
5X28
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2C
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(5) | | Descriptor: | 1,2-ETHANEDIOL, 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, ... | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2F
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
6JX0
 
 | |
5X27
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(5) | | Descriptor: | 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
6JWL
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with AZD9291 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Yun, C.H, Zhu, S.J, Yan, X.E. | | Deposit date: | 2019-04-21 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Basis of AZD9291 Selectivity for EGFR T790M.
J.Med.Chem., 63, 2020
|
|
5X26
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(3) | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-9-propan-2-yl-purine-2,8-diamine | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
8E3X
 
 | | Cryo-EM structure of the PAC1R-PACAP27-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
8E3Z
 
 | | Cryo-EM structure of the VPAC1R-VIP-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
8E3Y
 
 | | Cryo-EM structure of the VPAC1R-PACAP27-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
6P9X
 
 | | CRF1 Receptor Gs GPCR protein complex with CRF1 peptide | | Descriptor: | Corticoliberin, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-06-10 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation.
Mol.Cell, 77, 2020
|
|
6P9Y
 
 | | PAC1 GPCR Receptor complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-06-10 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation.
Mol.Cell, 77, 2020
|
|
