3J9I
 
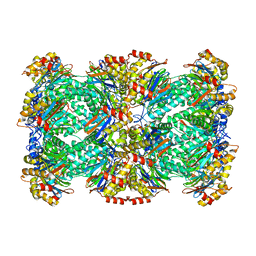 | | Thermoplasma acidophilum 20S proteasome | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Li, X, Mooney, P, Zheng, S, Booth, C, Braunfeld, M.B, Gubbens, S, Agard, D.A, Cheng, Y. | | Deposit date: | 2015-02-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Nat.Methods, 10, 2013
|
|
6KTR
 
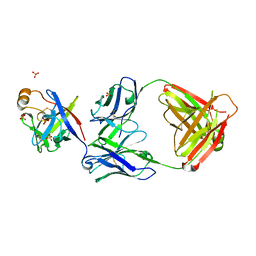 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
6AQJ
 
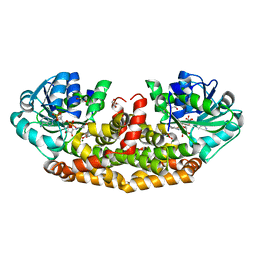 | | Crystal structures of Staphylococcus aureus ketol-acid reductoisomerase in complex with two transition state analogs that have biocidal activity. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Gracia, M, Lv, Y, Schembri, M.A, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.373 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
6BUL
 
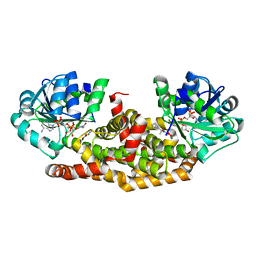 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 2 | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McFeary, R.P. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
6C5N
 
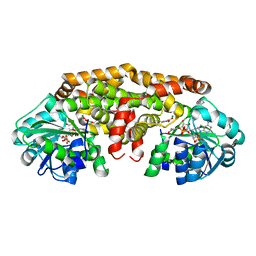 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | Descriptor: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
6C55
 
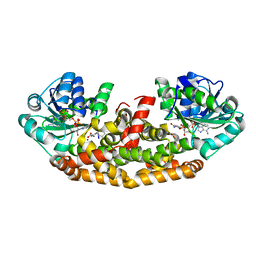 | | Crystal structure of Staphylococcus aureus Ketol-acid Reductosimerrase with hydroxyoxamate inhibitor 3 | | Descriptor: | (cyclohexylamino)(oxo)acetic acid, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schmbri, M, McGeary, R.P. | | Deposit date: | 2018-01-14 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
6D35
 
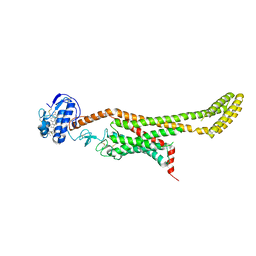 | | Crystal structure of Xenopus Smoothened in complex with cholesterol | | Descriptor: | CHOLESTEROL, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6D32
 
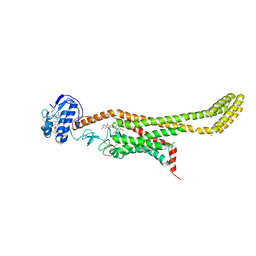 | | Crystal structure of Xenopus Smoothened in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
5HK1
 
 | | Human sigma-1 receptor bound to PD144418 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-(4-methylphenyl)-5-(1-propyl-3,6-dihydro-2H-pyridin-5-yl)-1,2-oxazole, SULFATE ION, ... | | Authors: | Schmidt, H.R, Zheng, S, Gurpinar, E, Koehl, A, Manglik, A, Kruse, A.C. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5051 Å) | | Cite: | Crystal structure of the human sigma 1 receptor.
Nature, 532, 2016
|
|
6U66
 
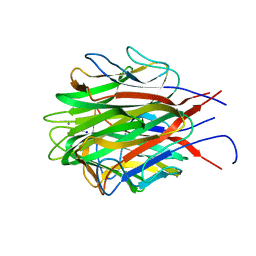 | | Structure of the trimeric globular domain of Adiponectin | | Descriptor: | Adiponectin, CALCIUM ION, SODIUM ION | | Authors: | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | Deposit date: | 2019-08-29 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
6U6N
 
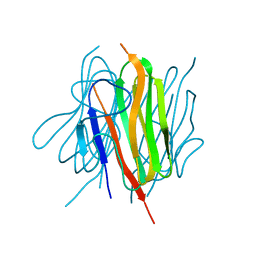 | | Structure of the trimeric globular domain of Adiponectin mutant - D187A Q188A | | Descriptor: | Adiponectin, CHLORIDE ION | | Authors: | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
3TAD
 
 | | Crystal Structure of the Liprin-alpha/Liprin-beta complex | | Descriptor: | GLYCEROL, Liprin-alpha-2, Liprin-beta-1 | | Authors: | Wei, Z, Zheng, S, Yu, C, Zhang, M. | | Deposit date: | 2011-08-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Liprin-mediated large signaling complex organization revealed by the liprin-alpha/CASK and liprin-alpha/liprin-beta complex structures
Mol.Cell, 43, 2011
|
|
3TAC
 
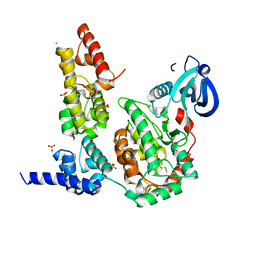 | | Crystal Structure of the Liprin-alpha/CASK complex | | Descriptor: | CHLORIDE ION, Liprin-alpha-2, Peripheral plasma membrane protein CASK, ... | | Authors: | Wei, Z, Zheng, S, Yu, C, Zhang, M. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Liprin-mediated large signaling complex organization revealed by the liprin-alpha/CASK and liprin-alpha/liprin-beta complex structures
Mol.Cell, 43, 2011
|
|
7F24
 
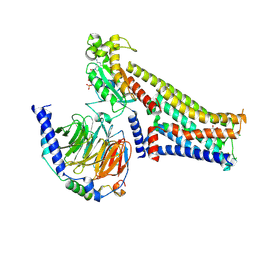 | |
7F0T
 
 | |
7F23
 
 | |
7F1O
 
 | |
7F1Z
 
 | |
4ZLK
 
 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
5WXM
 
 | | Crystal structure of the Imp3 and Mpp10 complex | | Descriptor: | SULFATE ION, U3 small nucleolar RNA-associated protein MPP10, U3 small nucleolar ribonucleoprotein protein IMP3 | | Authors: | Ye, K, Zheng, S. | | Deposit date: | 2017-01-07 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5YAX
 
 | | Crystal structure of a human neutralizing antibody bound to a HBV preS1 peptide | | Descriptor: | Large envelope protein, SODIUM ION, scFv1 antibody | | Authors: | Liu, X, Zheng, S, Ye, K, Sui, J. | | Deposit date: | 2017-09-02 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A potent human neutralizing antibody Fc-dependently reduces established HBV infections
Elife, 6, 2017
|
|
5WXL
 
 | | Crystal structure of the Rrs1 and Rpf2 complex | | Descriptor: | Regulator of ribosome biosynthesis, Ribosome biogenesis protein RPF2 | | Authors: | Ye, K, Zheng, S. | | Deposit date: | 2017-01-07 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5W3K
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex NADPH, Mg2+ and CPD | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Kandale, A, Schembri, M, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
8ZSZ
 
 | |
8ZSB
 
 | |
