4HFZ
 
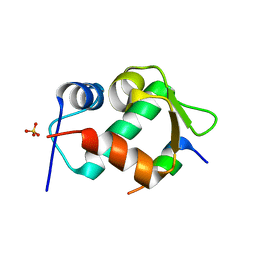 | | Crystal Structure of an MDM2/P53 Peptide Complex | | Descriptor: | Cellular tumor antigen p53, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Anil, B, Riedinger, C, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4QNL
 
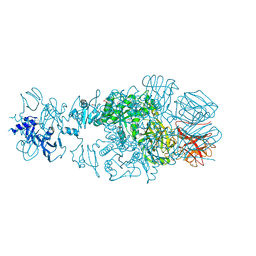 | | Crystal structure of tail fiber protein gp63.1 from E. coli phage G7C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Riccio, C, Browning, C, Prokhorov, N, Letarov, A, Leiman, P.G. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Function of bacteriophage G7C esterase tailspike in host cell adsorption.
Mol. Microbiol., 105, 2017
|
|
4HLQ
 
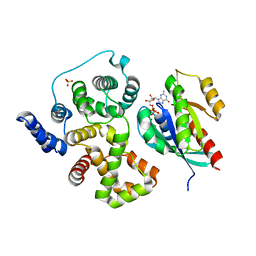 | | Crystal structure of human rab1b bound to GDP and BEF3 in complex with the GAP domain of TBC1D20 from homo sapiens | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gazdag, E.M, Gavriljuk, K, Itzen, A, Koetting, C, Gerwert, K, Goody, R.S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Catalytic mechanism of a mammalian Rab-RabGAP complex in atomic detail.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2V9J
 
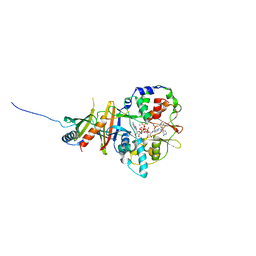 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with Mg.ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-23 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2O98
 
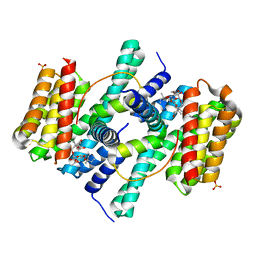 | | Structure of the 14-3-3 / H+-ATPase plant complex | | Descriptor: | 14-3-3-like protein C, FUSICOCCIN, Plasma membrane H+ ATPase, ... | | Authors: | Ottmann, C, Weyand, M, Wittinghofer, A, Oecking, C. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy
Mol.Cell, 25, 2007
|
|
1SO7
 
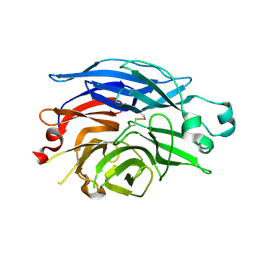 | | Maltose-induced structure of the human cytolsolic sialidase Neu2 | | Descriptor: | CHLORIDE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
2KV7
 
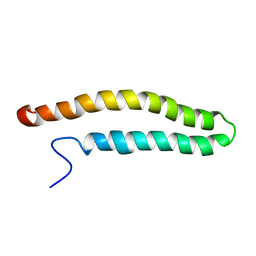 | | NMR solution structure of a soluble PrgI mutant from Salmonella Typhimurium | | Descriptor: | Protein prgI | | Authors: | Schmidt, H, Poyraz, O, Seidel, K, Delissen, F, Ader, C, Tenenboim, H, Goosmann, C, Laube, B, Thuenemann, A.F, Zychlinski, A, Baldus, M, Lange, A, Griesinger, C, Kolbe, M. | | Deposit date: | 2010-03-09 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein refolding is required for assembly of the type three secretion needle.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5NWX
 
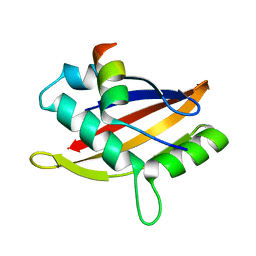 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
5NZK
 
 | | Crystal structure of UDP-glucose pyrophosphorylase from Leishmania major in complex with phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, UDP-glucose pyrophosphorylase | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
2L8I
 
 | | A biocompatible backbone modification? - Structure and dynamics of a triazole-linked DNA duplex | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G*(2L8)P*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*AP*AP*CP*GP*TP*CP*G)-3') | | Authors: | El-Sagheer, A, Brown, T, Ernsting, N, Dehmel, L, Griesinger, C, Mugge, C. | | Deposit date: | 2011-01-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Triazole-Linked DNA: Biocompatibility Explained.
Chemistry, 17, 2011
|
|
2LSV
 
 | | The NMR high resolution structure of yeast Tah1 in complex with the Hsp90 C-terminal tail | | Descriptor: | ATP-dependent molecular chaperone HSP82, TPR repeat-containing protein associated with Hsp90 | | Authors: | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
4OYR
 
 | | Competition of the small inhibitor PT91 with large fatty acyl substrate of the Mycobacterium tuberculosis enoyl-ACP reductase InhA by induced substrate-binding loop refolding | | Descriptor: | 2-(2-chloranylphenoxy)-5-hexyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2995 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
7DVO
 
 | | Structure of Reaction Intermediate of Cytochrome P450 NO Reductase (P450nor) Determined by XFEL | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Nomura, T, Kimura, T, Kanematsu, Y, Yamashita, K, Hirata, K, Ueno, G, Murakami, H, Hisano, T, Yamagiwa, R, Takeda, H, Gopalasingam, C, Yuki, K, Kousaka, R, Yanagasawa, S, Shoji, O, Kumasaka, T, Takano, Y, Ago, H, Yamamoto, M, Sugimoto, H, Tosha, T, Kubo, M, Shiro, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-lived intermediate in N 2 O generation by P450 NO reductase captured by time-resolved IR spectroscopy and XFEL crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5NKV
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 at pH 9.0 and 293 K. | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5NZJ
 
 | | Crystal structure of UDP-glucose pyrophosphorylase G45Y mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-glucose pyrophosphorylase, ... | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
5NZH
 
 | | Crystal structure of UDP-glucose pyrophosphorylase V402W mutant from Leishmania major | | Descriptor: | UDP-glucose pyrophosphorylase | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
5NZI
 
 | | Crystal structure of UDP-glucose pyrophosphorylase S374F mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
4OXN
 
 | | Substrate-like binding mode of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2926 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
2JK4
 
 | | Structure of the human voltage-dependent anion channel | | Descriptor: | VOLTAGE-DEPENDENT ANION-SELECTIVE CHANNEL PROTEIN 1 | | Authors: | Bayrhuber, M, Meins, T, Habeck, M, Becker, S, Giller, K, Villinger, S, Vonrhein, C, Griesinger, C, Zweckstetter, M, Zeth, K. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of the Human Voltage-Dependent Anion Channel.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5O4E
 
 | | Crystal structure of VEGF in complex with heterodimeric Fcab JanusCT6 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mlynek, G, Lobner, E, Kubinger, K, Humm, A, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
2K39
 
 | | Recognition dynamics up to microseconds revealed from RDC derived ubiquitin ensemble in solution | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Fares, C, Schroder, G, Walter, K, Becker, S, Meiler, J, Grubmuller, H, Griesinger, C, de Groot, B.L. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution.
Science, 320, 2008
|
|
5NZG
 
 | | Crystal structure of UDP-glucose pyrophosphorylase S374W mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
5NZM
 
 | | Crystal structure of UDP-glucose pyrophosphorylase from Leishmania major in complex with murrayamine-I | | Descriptor: | Murrayamine-I, UDP-glucose pyrophosphorylase | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
5NZL
 
 | | Crystal structure of UDP-glucose pyrophosphorylase from Leishmania major in complex with resveratrol | | Descriptor: | 1,2-ETHANEDIOL, RESVERATROL, UDP-glucose pyrophosphorylase | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
