4GU0
 
 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
3U0J
 
 | |
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | Authors: | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8XK2
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | Spike protein S1, VHH60 nanobody | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8XKI
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
4H0N
 
 | | Crystal structure of Spodoptera frugiperda DNMT2 E260A/E261A/K263A mutant | | Descriptor: | CALCIUM ION, DNMT2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Li, S, Du, J, Yang, H, Yin, J, Zhong, J, Ding, J. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Functional and structural characterization of DNMT2 from Spodoptera frugiperda.
J Mol Cell Biol, 5, 2013
|
|
9J6O
 
 | | Crystal Structure of bromodomain of human CBP in complex with the inhibitor CZL-046 | | Descriptor: | (3~{S},5~{S})-1-[3,4-bis(fluoranyl)phenyl]-5-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]-3-fluoranyl-pyrrolidin-2-one, CHLORIDE ION, CREB-binding protein | | Authors: | Zhang, Y, Zhang, C, Chen, Z, Yang, H, Huang, X, Li, Y, Xu, Y. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of CZL-046 with an ( S )-3-Fluoropyrrolidin-2-one Scaffold as a p300 Bromodomain Inhibitor for the Treatment of Multiple Myeloma.
J.Med.Chem., 67, 2024
|
|
4KTH
 
 | | Structure of A/Hubei/1/2010 H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Shore, D.A, Yang, H, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Antigenic Variation among Diverse Clade 2 H5N1 Viruses.
Plos One, 8, 2013
|
|
8ZVX
 
 | | Crystal structure of snFPITE-n2 | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Chen, X, Yang, H. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A facultative plasminogen-independent thrombolytic enzyme from Sipunculus nudus.
Nat Commun, 16, 2025
|
|
8ZVS
 
 | | Crystal structure of snFPITE-n1 | | Descriptor: | SULFATE ION, phenylmethanesulfonic acid, snFPITE-n1 | | Authors: | Chen, X, Yang, H, Hu, Y.L. | | Deposit date: | 2024-06-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | A facultative plasminogen-independent thrombolytic enzyme from Sipunculus nudus.
Nat Commun, 16, 2025
|
|
4KW1
 
 | | Structure of a/egypt/n03072/2010 h5 ha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Shore, D.A, Yang, H, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and antigenic variation among diverse clade 2 H5N1 viruses.
Plos One, 8, 2013
|
|
1WNO
 
 | | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Hu, H, Wang, G, Yang, H, Zhou, J, Mo, L, Yang, K, Jin, C, Jin, C, Rao, Z. | | Deposit date: | 2004-08-07 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407
To be Published
|
|
7VUX
 
 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8W6C
 
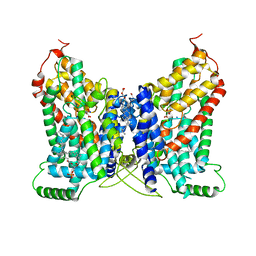 | | CryoEM structure of NaDC1 with Citrate | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CHOLESTEROL HEMISUCCINATE, CITRIC ACID, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Dai, L, Zhang, Y, Shen, Y, Chen, Y, Shi, T, Yang, H, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8GXH
 
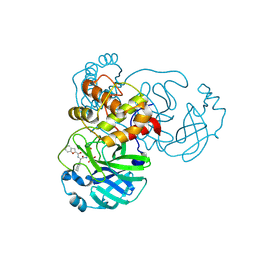 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8WDD
 
 | | Crystal structure of BSA in complex with B1 | | Descriptor: | Albumin, ~{N},~{N}-dimethyl-6-[(~{E})-2-(1-methylpyridin-1-ium-4-yl)ethenyl]naphthalen-2-amine | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of BSA in complex with B1
To Be Published
|
|
8KFO
 
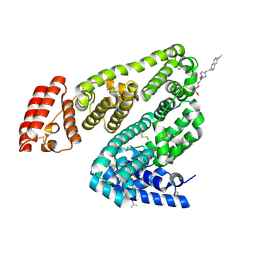 | | Crystal structure of BSA in complex with B3 | | Descriptor: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Albumins constrainting the conformation of mitochondria-targeted photosensitizers for tumor-specific photodynamic therapy.
Biomaterials, 315, 2025
|
|
7VAH
 
 | |
7WIX
 
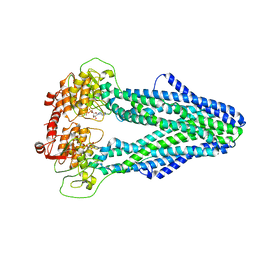 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
