6P9M
 
 | |
5TW3
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2016-11-11 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural and Preclinical Studies of Computationally Designed Non-Nucleoside Reverse Transcriptase Inhibitors for Treating HIV infection.
Mol. Pharmacol., 91, 2017
|
|
7NRP
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3'), RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-03-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OOO
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG containing an LNA-Amide-LNA modification | | Descriptor: | DNA (5'-D(*CP*TP*(05A)P*TP*CP*TP*TP*TP*G)-3'), MAGNESIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OOS
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG | | Descriptor: | DNA (5'-D(*CP*TP*(05K)P*TP*CP*TP*TP*TP*G)-3'), RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), STRONTIUM ION | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OZZ
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG with LNA-amide modification | | Descriptor: | DNA (5'-D(*CP*TP*(05H)P*TP*CP*TP*TP*TP*G)-3'), POTASSIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-06-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
5VND
 
 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | Authors: | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | Deposit date: | 2017-04-30 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
6OE3
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Kudalkar, S.N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and pharmacological evaluation of a novel non-nucleoside reverse transcriptase inhibitor as a promising long acting nanoformulation for treating HIV.
Antiviral Res., 167, 2019
|
|
4PTN
 
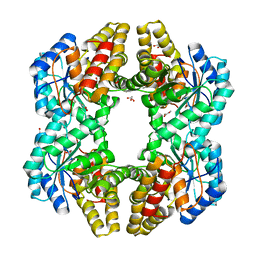 | | Crystal Structure of YagE, a KDG aldolase protein in complex with Magnesium cation coordinated L-glyceraldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-glyceraldehyde, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12.
Proteins, 71, 2008
|
|
7RQ6
 
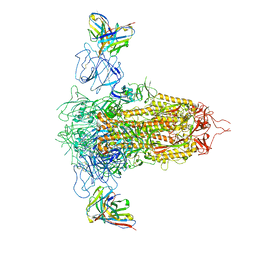 | | Cryo-EM structure of SARS-CoV-2 spike in complex with non-neutralizing NTD-directed CV3-13 Fab isolated from convalescent individual | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CV3-13 Fab heavy chain, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-08-05 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection.
Cell Rep, 38, 2022
|
|
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
7Q6Z
 
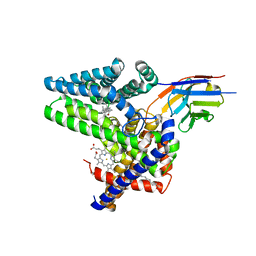 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to IMP-1575 | | Descriptor: | 2-(2-methylpropylamino)-1-[(4R)-4-(6-methylpyridin-2-yl)-6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl]ethanone, CHOLESTEROL, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
7Q1U
 
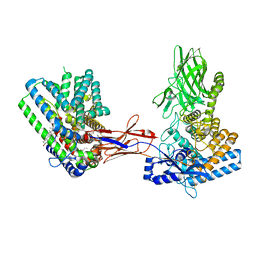 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Composite Map) | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
8VBB
 
 | |
8VBA
 
 | |
7NAB
 
 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
6BMM
 
 | | Structure of human DHHC20 palmitoyltransferase, space group P21 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,5S)-hexane-2,5-diol, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
7UL1
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
6DGB
 
 | | Crystal structure of the C-terminal catalytic domain of IS1535 TnpA, an IS607-like serine recombinase | | Descriptor: | IS607 family transposase IS1535 | | Authors: | Chen, W.Y, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
9JP7
 
 | |
9JP5
 
 | |
3MIO
 
 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis at pH 6.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-11 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
