6UX2
 
 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7AS0
 
 | | Influenza A PB2 in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, BROMIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS1
 
 | | Influenza A PB2 (F404Y mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, CHLORIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS2
 
 | | Influenza A PB2 (M431 mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS3
 
 | | Influenza A PB2 (H357N mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
6MVQ
 
 | | HCV NS5B 1b N316 bound to Compound 31 | | Descriptor: | (4-{1-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-1H-1,2,4-triazol-5-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
6QBG
 
 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 14 | | Descriptor: | (3~{S},7~{S},8~{S})-8-(naphthalen-2-ylmethyl)-7-oxidanyl-3-propan-2-yl-1,4,9-triazacyclohenicosane-2,5,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
8AB5
 
 | | Structure of E. coli GlpG in complex with peptide derived inhibitor Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl] | | Descriptor: | Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl], Rhomboid protease GlpG | | Authors: | Skerlova, J, Polovinkin, V, Bach, K, Borshchevskiy, V, Strisovsky, K. | | Deposit date: | 2022-07-04 | | Release date: | 2023-10-25 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extensive targeting of chemical space at the prime side of ketoamide inhibitors of rhomboid proteases by branched substituents empowers their selectivity and potency.
Eur.J.Med.Chem., 275, 2024
|
|
6QBH
 
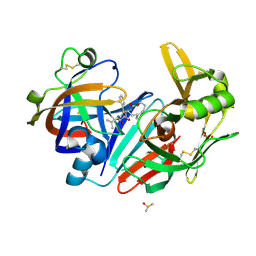 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 33 | | Descriptor: | (4~{S},5~{S},9~{S})-5-oxidanyl-4-(phenylmethyl)-9-propan-2-yl-1-oxa-3,8,11-triazacyclodocosane-2,7,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
6QCB
 
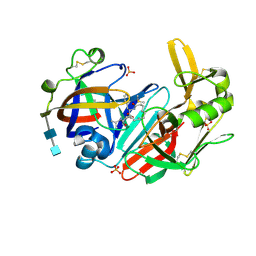 | | Crystal structure of human cathepsin D in complex with macrocyclic inhibitor 9 | | Descriptor: | (3~{S},7~{S},8~{S})-7-oxidanyl-8-(phenylmethyl)-3-propan-2-yl-1,4,9-triazacyclohenicosane-2,5,10-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin D, ... | | Authors: | Brynda, J, Houstecka, R, Majer, P, Mares, M. | | Deposit date: | 2018-12-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomimetic Macrocyclic Inhibitors of Human Cathepsin D: Structure-Activity Relationship and Binding Mode Analysis.
J.Med.Chem., 63, 2020
|
|
7XZO
 
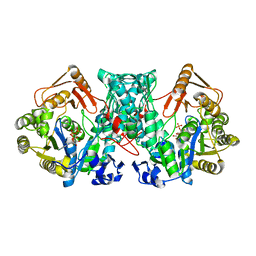 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
7XZN
 
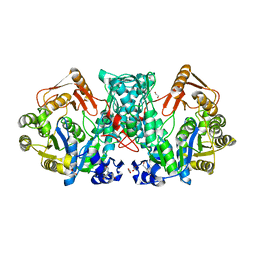 | |
7XZP
 
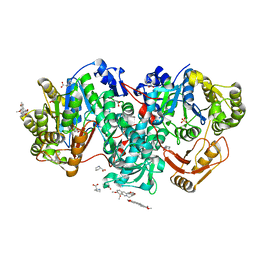 | |
3ELM
 
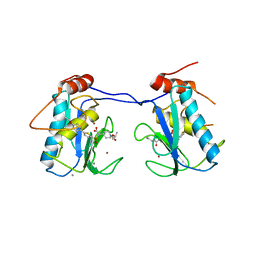 | | Crystal Structure of MMP-13 Complexed with Inhibitor 24f | | Descriptor: | (2R)-({[5-(4-ethoxyphenyl)thiophen-2-yl]sulfonyl}amino){1-[(1-methylethoxy)carbonyl]piperidin-4-yl}ethanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Kulathila, R, Monovich, L, Koehn, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent, selective, and orally active carboxylic acid based inhibitors of matrix metalloproteinase-13
J.Med.Chem., 52, 2009
|
|
3ZXH
 
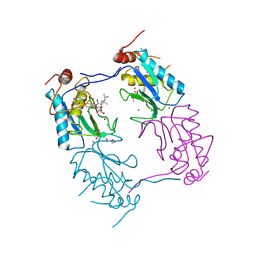 | |
3B3O
 
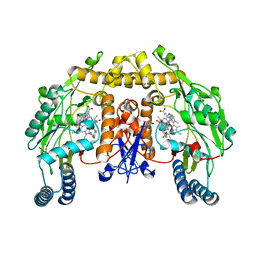 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
6MVP
 
 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}phenyl)boronic acid, Genome polyprotein | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
6MVK
 
 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{(4S)-3-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-2-oxo-1,3-oxazolidin-4-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
6W8B
 
 | | Structure of DNMT3A in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
3DQS
 
 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
6W8D
 
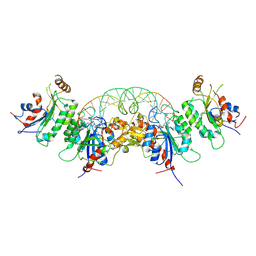 | | Structure of DNMT3A (R882H) in complex with CGT DNA | | Descriptor: | CGT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W8J
 
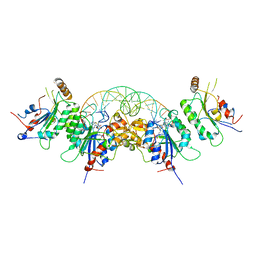 | | Structure of DNMT3A (R882H) in complex with CAG DNA | | Descriptor: | CAG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W89
 
 | | Structure of DNMT3A (R882H) in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), CITRIC ACID, DNA (cytosine-5)-methyltransferase 3-like, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
3DQT
 
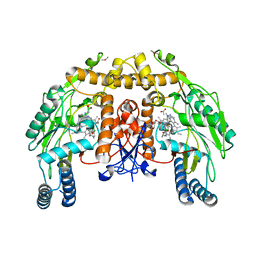 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{trans-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
