6Y20
 
 | |
6Z10
 
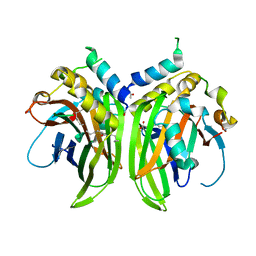
 | | Crystal structure of a humanized (K18E, K269N) rat succinate receptor SUCNR1 (GPR91) in complex with a nanobody and antagonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[[3-[4-chloranyl-2-fluoranyl-5-[(3~{R})-piperidin-3-yl]oxy-phenyl]-2-fluoranyl-phenyl]carbonylamino]-5-fluoranyl-phenyl]ethanoic acid, CHLORIDE ION, ... | | Authors: | Haffke, M, Villard, F. | | Deposit date: | 2020-05-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Discovery and Optimization of Novel SUCNR1 Inhibitors: Design of Zwitterionic Derivatives with a Salt Bridge for the Improvement of Oral Exposure.
J.Med.Chem., 63, 2020
|
|
6SEO
 
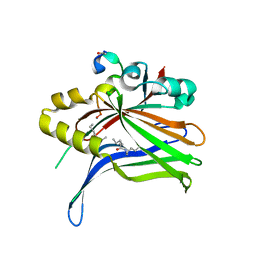 | | TEAD4 bound to a FAM181B peptide | | Descriptor: | Protein FAM181B, Transcriptional enhancer factor TEF-3 | | Authors: | Scheufler, C, Villard, F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors.
Protein Sci., 29, 2020
|
|
6SEN
 
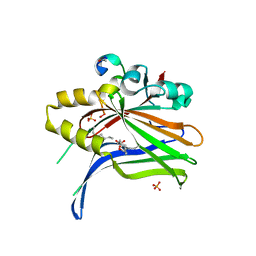 | | TEAD4 bound to a FAM181A peptide | | Descriptor: | Protein FAM181A, SULFATE ION, Transcriptional enhancer factor TEF-3 | | Authors: | Scheufler, C, Villard, F. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors.
Protein Sci., 29, 2020
|
|
5JMY
 
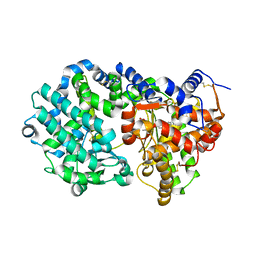 | | NEPRILYSIN COMPLEXED WITH LBQ657 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Schiering, N, Wiesmann, C. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of neprilysin in complex with the active metabolite of sacubitril.
Sci Rep, 6, 2016
|
|
8C17
 
 | | Crystal structure of TEAD4 in complex with peptide 1 | | Descriptor: | GLYCEROL, MYRISTIC ACID, Stapled peptide, ... | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and Structural Characterization of a Peptidic Inhibitor of the YAP:TEAD Interaction That Binds to the alpha-Helix Pocket on TEAD.
Acs Chem.Biol., 18, 2023
|
|
3V4L
 
 | |
3V55
 
 | | Human MALT1 (334-719) in its ligand free form | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M, Wiesmann, C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Determinants of MALT1 Protease Activity.
J.Mol.Biol., 419, 2012
|
|
4A92
 
 | | Full-length HCV NS3-4A protease-helicase in complex with a macrocyclic protease inhibitor. | | Descriptor: | (1'R,2R,2'S,6S,24AS)-17-FLUORO-6-(1-METHYL-2-OXOPIPERIDINE-3-CARBOXAMIDO)-19,19-DIOXIDO-5,21,24-TRIOXO-2'-VINYL-1,2,3,5,6,7,8,9,10,11,12,13,14,20,21,23,24,24A-OCTADECAHYDROSPIRO[BENZO[S]PYRROLO[2,1-G][1,2,5,8,18]THIATETRAAZACYCLOICOSINE-22,1'-CYCLOPRO-2-CARBOXYLATEPAN]-2-YL 4-FLUOROISOINDOLINE, SERINE PROTEASE NS3, ZINC ION | | Authors: | Schiering, N, D'Arcy, A, Simic, O, Eder, J, Raman, P, Svergun, D.I, Bodendorf, U. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Macrocyclic Hcv Ns3/4A Protease Inhibitor Interacts with Protease and Helicase Residues in the Complex with its Full- Length Target.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3V4O
 
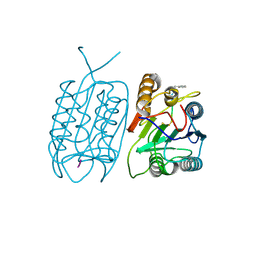 | |
6YN9
 
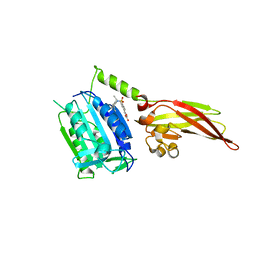 | | MALT1(329-728) in complex with a sulfonamide containing compound | | Descriptor: | 5-[4-[(2,6-diethylphenyl)sulfamoyl]-3-methyl-phenyl]furan-3-carboxylic acid, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
6YN8
 
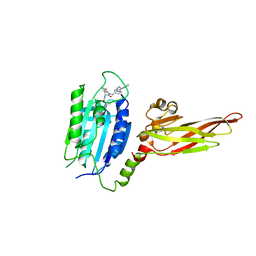 | | Human MALT1(334-719) in complex with a tetrazole containing compound | | Descriptor: | 3-azanyl-3-methyl-~{N}-[(3~{R})-4-oxidanylidene-5-[[4-[2-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]-2,3-dihydro-1,5-benzoxazepin-3-yl]butanamide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
6THP
 
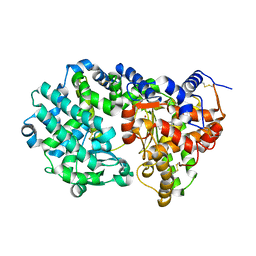 | |
6FFS
 
 | | Structure-based design and synthesis of macrocyclic human rhinovirus 3C protease inhibitors | | Descriptor: | 3C Protease, SULFATE ION, ~{N}-[(2~{S},5~{S},14~{S})-2-[(4-fluorophenyl)methyl]-5-(hydroxymethyl)-9-methyl-3,8,15-tris(oxidanylidene)-1,4,9-triazacyclopentadec-14-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wiesmann, C, Farady, C. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design and synthesis of macrocyclic human rhinovirus 3C protease inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6F7I
 
 | | human MALT1(329-728) IN COMPLEX WITH MLT-747 | | Descriptor: | 1-[2-chloranyl-7-[(1~{S})-1-methoxyethyl]pyrazolo[1,5-a]pyrimidin-6-yl]-3-(5-chloranyl-6-pyrrolidin-1-ylcarbonyl-pyridin-3-yl)urea, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Renatus, M, Renatus, M. | | Deposit date: | 2017-12-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient.
Nat. Chem. Biol., 15, 2019
|
|
8PFP
 
 | |
6FFN
 
 | |
6H4A
 
 | | Human MALT1(329-728) in complex with MLT-748 | | Descriptor: | 1-[2-chloranyl-7-[(1~{R},2~{R})-1,2-dimethoxypropyl]pyrazolo[1,5-a]pyrimidin-6-yl]-3-[5-chloranyl-6-(1,2,3-triazol-2-yl)pyridin-3-yl]urea, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M, Renatus, M. | | Deposit date: | 2018-07-20 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient.
Nat. Chem. Biol., 15, 2019
|
|
8A8Q
 
 | |
8P0M
 
 | | Crystal structure of TEAD3 in complex with IAG933 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-[(2~{S})-5-chloranyl-6-fluoranyl-2-phenyl-2-[(2~{S})-pyrrolidin-2-yl]-3~{H}-1-benzofuran-4-yl]-5-fluoranyl-6-(2-hydroxyethyloxy)-~{N}-methyl-pyridine-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scheufler, C, Villard, F, Chau, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.
Nat Cancer, 5, 2024
|
|
6R8X
 
 | |
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7QFM
 
 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7Z6U
 
 | | Pim1 in complex with (E)-4-((6-amino-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(6-azanyl-2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Isoform 1 of Serine/threonine-protein kinase pim-1, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
8AFR
 
 | | Pim1 in complex with 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid and Pimtide | | Descriptor: | 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-07-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
