2WO5
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form I | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNN
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, SODIUM ION | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNZ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 crystal form I | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-ETHOXYETHANOL, LACTIC ACID, ... | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNQ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in space group P21 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WKJ
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate at 1.45A resolution in space group P212121 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, PYRUVIC ACID | | Authors: | Campeotto, I, Carr, S.B, Trinh, C.H, Nelson, A.S, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2009-06-11 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of an Escherichia coli N-acetyl-D-neuraminic acid lyase mutant, E192N, in complex with pyruvate at 1.45 angstrom resolution.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 65, 2009
|
|
5LKY
 
 | | X-ray crystal structure of N-acetylneuraminic acid lyase in complex with pyruvate, with the phenylalanine at position 190 replaced with the non-canonical amino acid dihydroxypropylcysteine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetylneuraminate lyase | | Authors: | Windle, C.L, Trinh, C.H, Pearson, A.R, Nelson, A.S, Berry, A. | | Deposit date: | 2016-07-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending enzyme molecular recognition with an expanded amino acid alphabet.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4AMA
 
 | | Crystal Structure of N-acetylneuraminic acid lyase from Staphylococcus aureus with the chemical modification thia-lysine at position 165 in complex with pyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Pearson, A.R, Berry, A. | | Deposit date: | 2012-03-08 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the recovery of aldolase activity in N-acetylneuraminic acid lyase by replacement of the catalytically active lysine with gamma-thialysine by using a chemical mutagenesis strategy.
Chembiochem, 14, 2013
|
|
5A8G
 
 | | Crystal structure of the wild-type Staphylococcus aureus N- acetylneurminic acid lyase in complex with fluoropyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Stockwell, J, Daniels, A.D, Windle, C.L, Harman, T, Woodhall, T, Trinh, C.H, Lebel, T, Pearson, A.R, Mulholland, K, Berry, A, Nelson, A. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evaluation of Fluoropyruvate as Nucleophile in Reactions Catalysed by N-Acetyl Neuraminic Acid Lyase Variants: Scope, Limitations and Stereoselectivity.
Org.Biomol.Chem., 14, 2016
|
|
6HNM
 
 | | Crystal structure of IdmH 96-104 loop truncation variant | | Descriptor: | putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
6HNN
 
 | | Crystal structure of wild-type IdmH, a putative polyketide cyclase from Streptomyces antibioticus | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
6HNL
 
 | | Selenomethionine derivative of IdmH 96-104 loop truncation variant | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
1GYN
 
 | | Class II fructose 1,6-bisphosphate aldolase with Cadmium (not Zinc) in the active site | | Descriptor: | CADMIUM ION, FRUCTOSE-BISPHOSPHATE ALDOLASE II | | Authors: | Hall, D.R, Kemp, L.E, Leonard, G.A, Berry, A, Hunter, W.N. | | Deposit date: | 2002-04-27 | | Release date: | 2003-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Organization of Divalent Cations in the Active Site of Cadmium Escherichia Coli Fructose 1,6-Bisphosphate Aldolase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4AHP
 
 | | Crystal Structure of Wild Type N-acetylneuraminic acid lyase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
2WPB
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate and the inhibitor (2R,3R)-2,3,4- trihydroxy-N,N-dipropylbutanamide in space group P21 crystal form I | | Descriptor: | (2R,3R)-2,3,4-TRIHYDROXY-N,N-DIPROPYLBUTANAMIDE, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2XFW
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate in crystal form III | | Descriptor: | N-ACETYLNEURAMINIC ACID LYASE, PENTAETHYLENE GLYCOL, PYRUVIC ACID | | Authors: | Campeotto, I, Murshudov, G.N, Bolt, A.H, Trinh, C.H, Phillips, S.E.V, Nelson, A, Pearson, A.R, Berry, A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
To be Published
|
|
4AHQ
 
 | | Crystal Structure of N-acetylneuraminic acid lyase mutant K165C from Staphylococcus aureus | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
4AHO
 
 | | Crystal Structure of N-acetylneuraminic acid lyase from Staphylococcus aureus with the chemical modification thia-lysine at position 165 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
4AH7
 
 | | Structure of Wild Type Stapylococcus aureus N-acetylneuraminic acid lyase in complex with pyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Poyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Pearson, A.R, Berry, A. | | Deposit date: | 2012-02-03 | | Release date: | 2013-01-23 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
4BWL
 
 | | Structure of the Y137A mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate, N-acetyl-D-mannosamine and N- acetylneuraminic acid | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, N-ACETYLNEURAMINATE LYASE, ... | | Authors: | Campeotto, I, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2013-07-03 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, Qm/Mm Simulation and Mutagenesis.
Acs Chem.Biol., 9, 2014
|
|
2XCT
 
 | | The twinned 3.35A structure of S. aureus Gyrase complex with Ciprofloxacin and DNA | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 5'-D(AP*GP*CP*CP*GP*TP*AP*G)-3', 5'-D(GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3', ... | | Authors: | Bax, B.D, Chan, P, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCR
 
 | | The 3.5A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase complexed with GSK299423 and DNA | | Descriptor: | 5'-D(*5UA*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCQ
 
 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCS
 
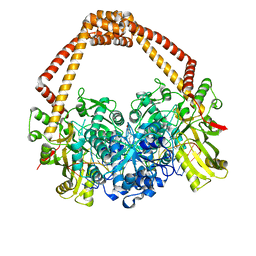 | | The 2.1A crystal structure of S. aureus Gyrase complex with GSK299423 and DNA | | Descriptor: | 5'-5UA*D(GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP *CP*TP*AP*CP*GP*GP*CP*T)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, DNA GYRASE SUBUNIT B, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCO
 
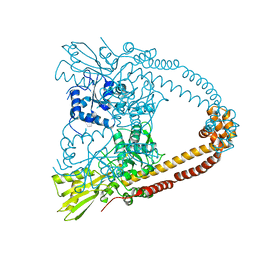 | | The 3.1A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | CALCIUM ION, DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2AIA
 
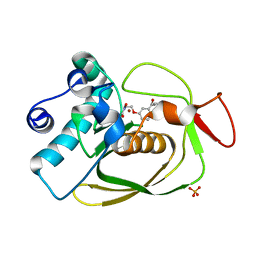 | | S.pneumoniae PDF complexed with SB-543668 | | Descriptor: | 2-(3-BENZOYLPHENOXY)ETHYL(HYDROXY)FORMAMIDE, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Smith, K.J, Petit, C.M, Aubart, K, Smyth, M, McManus, E, Jones, J, Fosberry, A, Lewis, C, Lonetto, M, Christensen, S.B. | | Deposit date: | 2005-07-29 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Variation and inhibitor binding in polypeptide deformylase from four different bacterial species.
Protein Sci., 12, 2003
|
|
