5MYV
 
 | | Crystal structure of SRPK2 in complex with compound 1 | | Descriptor: | 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, DIMETHYL SULFOXIDE, SRSF protein kinase 2,SRSF protein kinase 2, ... | | Authors: | Chaikuad, A, Pike, A.C.W, Savitsky, P, von Delft, F, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
5MP0
 
 | | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, m7GpppN-mRNA hydrolase | | Authors: | Mathea, S, Salah, E, Velupillai, S, Tallant, C, Pike, A.C.W, Bushell, S.R, Faust, B, Wang, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Huber, K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain
To Be Published
|
|
5NNC
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H3K9ac/K14ac) | | Descriptor: | Bromodomain-containing protein 4, Histone H3 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5NND
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H3K9ac/K14ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Histone H3 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
4AP2
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 2.8A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CULLIN-3, ... | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Filippakopoulos, P, Ayinampudi, V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
4APF
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 3.1A resolution | | Descriptor: | CULLIN 3, KELCH-LIKE PROTEIN 11 | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Vollmar, M, Ugochukwu, E, Muniz, J.R.C, Ayinampudi, V, Savitsky, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
4ASC
 
 | | Crystal structure of the Kelch domain of human KBTBD5 | | Descriptor: | 1,2-ETHANEDIOL, KELCH REPEAT AND BTB DOMAIN-CONTAINING PROTEIN 5 | | Authors: | Canning, P, Ayinampudi, V, Krojer, T, Strain-Damerell, C, Raynor, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
3MY0
 
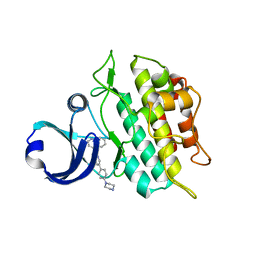 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | Authors: | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
3MTL
 
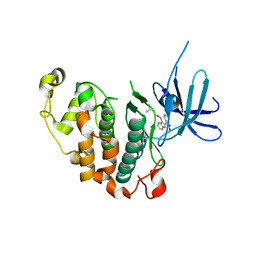 | | Crystal structure of the PCTAIRE1 kinase in complex with Indirubin E804 | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cell division protein kinase 16 | | Authors: | Krojer, T, Sharpe, T.D, Roos, A, Savitsky, P, Amos, A, Ayinampudi, V, Berridge, G, Fedorov, O, Keates, T, Phillips, C, Burgess-Brown, N, Zhang, Y, Pike, A.C.W, Muniz, J, Vollmar, M, Thangaratnarajah, C, Rellos, P, Ugochukwu, E, Filippakopoulos, P, Yue, W, Das, S, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16.
Biochem.J., 474, 2017
|
|
3QK3
 
 | | Crystal structure of human beta-crystallin B3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-crystallin B3, SULFATE ION | | Authors: | Krojer, T, Gileadi, C, Cocking, R, Muniz, J, Pilka, E, Yue, W.W, Vollmar, M, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: |
|
|
7AIO
 
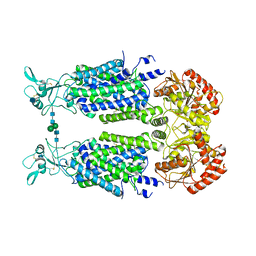 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Subclass) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
7AIN
 
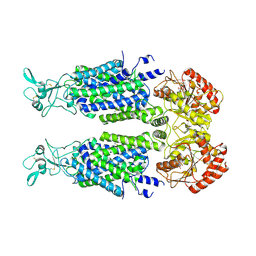 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
6FT3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{R})-cyclopropyl(oxidanyl)methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FSY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 3-[[4,4-bis(fluoranyl)piperidin-1-yl]methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FDZ
 
 | | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib | | Descriptor: | Momelotinib, Serine/threonine-protein kinase ULK3 | | Authors: | Mathea, S, Salah, E, Moroglu, M, Scorah, A, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Knapp, S. | | Deposit date: | 2017-12-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib
To Be Published
|
|
6GY5
 
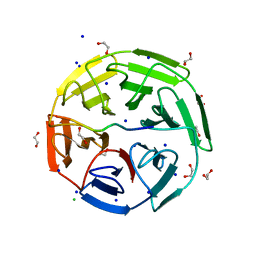 | | Crystal structure of the kelch domain of human KLHL20 in complex with DAPK1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Chen, Z, Hozjan, V, Strain-Damerell, C, Williams, E, Wang, D, Cooper, C.D.O, Sanvitale, C.E, Fairhead, M, Carpenter, E.P, Pike, A.C.W, Krojer, T, Srikannathasan, V, Sorrell, F, Johansson, C, Mathea, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Structural Basis for Recruitment of DAPK1 to the KLHL20 E3 Ligase.
Structure, 27, 2019
|
|
6G0R
 
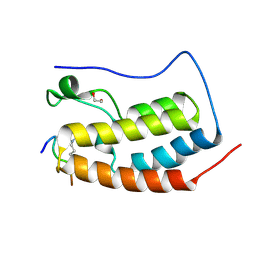 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated POLR2A peptide (K775ac/K778ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DNA-directed RNA polymerase II subunit RPB1 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0P
 
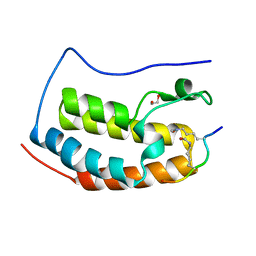 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated E2F1 peptide (K117ac/K120ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcription factor E2F1 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, Sorrell, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
3IQ7
 
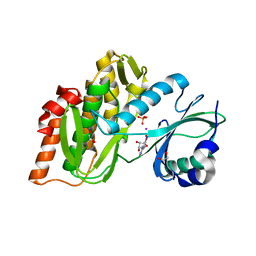 | | Crystal Structure of human Haspin in complex with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-19 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and functional characterization of the atypical human kinase haspin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IU6
 
 | | Crystal structure of the sixth bromodomain of human poly-bromodomain containing protein 1 (PB1) | | Descriptor: | 1,2-ETHANEDIOL, Protein polybromo-1, ZINC ION | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-30 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3K1Z
 
 | | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Haloacid dehalogenase-like hydrolase domain-containing protein 3 | | Authors: | Ugochukwu, E, Guo, K, Yue, W.W, Pilka, E, Picaud, S, Muniz, J, Pike, A.C.W, Krojer, T, Gomes, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-11-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3)
To be Published
|
|
3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
3K2L
 
 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) | | Descriptor: | CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2, SODIUM ION, ... | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Soundararajan, M, Krojer, T, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of Down Syndrome Kinases, DYRKs, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
