3J9L
 
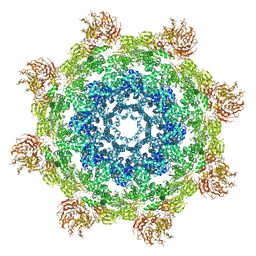 | | Structure of Dark apoptosome from Drosophila melanogaster | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
3JA7
 
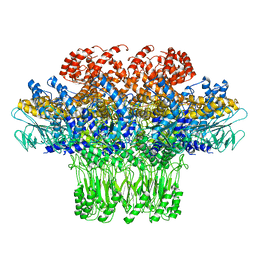 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
7RNN
 
 | | Human ASIC1a-Nb.C1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Nanobodies Nb.C1 | | Authors: | Wu, Y, Chen, Z, Sigworth, F.J, Canessa, C.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure and analysis of nanobody binding to the human ASIC1a ion channel.
Elife, 10, 2021
|
|
3KPQ
 
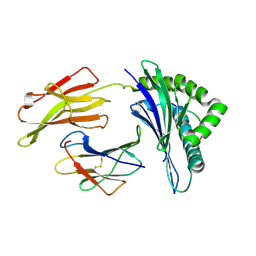 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
6LUQ
 
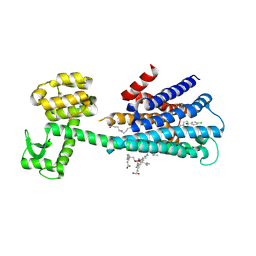 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
4MYW
 
 | | Structure of HSV-2 gD bound to nectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
6TN8
 
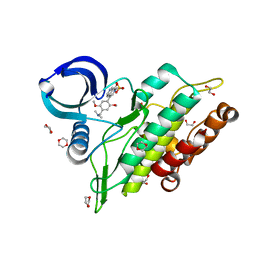 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound BI-9564 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, ... | | Authors: | Williams, E.P, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound BI-9564
To Be Published
|
|
8YUU
 
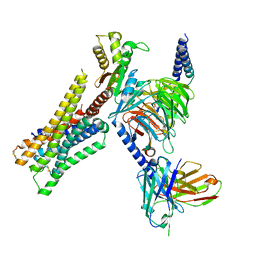 | | Cryo-EM structure of the histamine-bound H3R-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUV
 
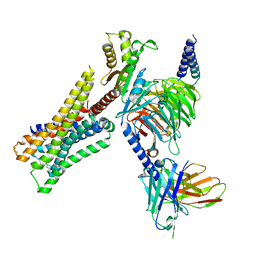 | | Cryo-EM structure of the immepip-bound H3R-Gi complex | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUT
 
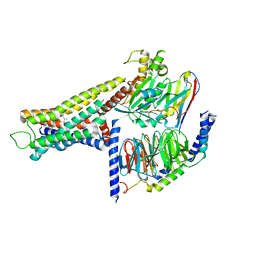 | | Cryo-EM structure of the amthamine-bound H2R-Gs complex | | Descriptor: | 5-(2-azanylethyl)-4-methyl-1,3-thiazol-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
6LTU
 
 | | Crystal structure of Cas12i2 ternary complex with double Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LTR
 
 | | Crystal structure of Cas12i2 ternary complex with single Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LU0
 
 | | Crystal structure of Cas12i2 ternary complex with 12 nt spacer | | Descriptor: | Cas12i2, DNA (5'-D(*GP*CP*CP*GP*CP*TP*TP*TP*CP*TP*T)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*CP*TP*CP*TP*GP*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*C)-3'), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-24 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LTP
 
 | | Crystal structure of Cas12i2 binary complex | | Descriptor: | Cas12i2, crRNA (56-mer RNA) | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
8DYO
 
 | | Cryo-EM structure of Importin-4 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-4, ... | | Authors: | Bernardes, N.E, Fung, H.Y.J, Li, Y, Chen, Z, Chook, Y.M. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1TQF
 
 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
5WRW
 
 | | Structure of human apo-SRP72 | | Descriptor: | SULFATE ION, Signal recognition particle subunit SRP72 | | Authors: | Gao, Y, Chen, Z. | | Deposit date: | 2016-12-04 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
8YPU
 
 | | Cryo-EM structure of ButCD complex | | Descriptor: | Membrane protein, SusC/RagA family TonB-linked outer membrane protein | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.
Nat Commun, 15, 2024
|
|
8YPT
 
 | | Cryo-EM structure of BfUbb-ButCD complex | | Descriptor: | Membrane protein, TonB-linked outer membrane protein, Ubiquitin-like protein | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.
Nat Commun, 15, 2024
|
|
4XOI
 
 | | Structure of hsAnillin bound with RhoA(Q63L) at 2.1 Angstroms resolution | | Descriptor: | Actin-binding protein anillin, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sun, L, Guan, R, Lee, I.-J, Liu, Y, Chen, M, Wang, J, Wu, J, Chen, Z. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
4XXB
 
 | | Crystal structure of human MDM2-RPL11 | | Descriptor: | 60S ribosomal protein L11, BETA-MERCAPTOETHANOL, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Zheng, J, Chen, Z. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Genes Dev., 29, 2015
|
|
5X0X
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
4O7X
 
 | |
4NRQ
 
 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
