7N9V
 
 | |
6BWY
 
 | | DNA substrate selection by APOBEC3G | | Descriptor: | DNA (30-MER), PHOSPHATE ION, Protection of telomeres protein 1, ... | | Authors: | Ziegler, S.J, Buzovetsky, O. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into DNA substrate selection by APOBEC3G from structural, biochemical, and functional studies.
PLoS ONE, 13, 2018
|
|
4WF6
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor MK-31 | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ... | | Authors: | Maize, K.M, De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2014-09-12 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6521 Å) | | Cite: | Probing the S2' Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates.
J.Med.Chem., 58, 2015
|
|
5V9L
 
 | |
5V9O
 
 | | KRAS G12C inhibitor | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C.
Cell Chem Biol, 24, 2017
|
|
5XHQ
 
 | | Apolipoprotein N-acyl Transferase | | Descriptor: | Apolipoprotein N-acyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yingzhi, X, Yong, X, Guangyuan, L, Fei, S. | | Deposit date: | 2017-04-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Crystal structure of E. coli apolipoprotein N-acyl transferase
Nat Commun, 8, 2017
|
|
5XDZ
 
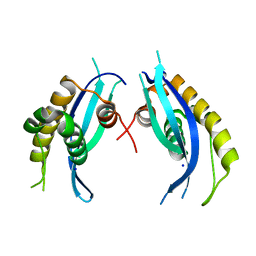 | | Crystal structure of zebrafish SNX25 PX domain | | Descriptor: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
7VWZ
 
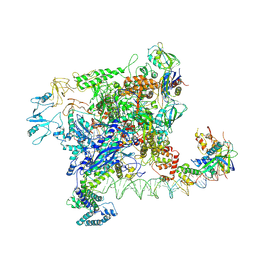 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
7VWY
 
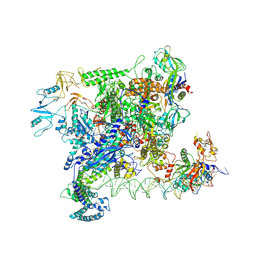 | |
5Y5W
 
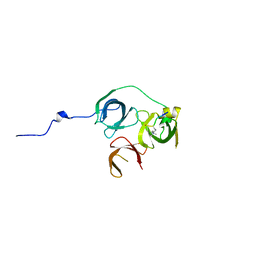 | |
6I8L
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8B
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8Y
 
 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
7CKF
 
 | | The N-terminus of interferon-inducible antiviral protein-dimer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Yang, H.T. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4HJX
 
 | | Crystal structure of F2YRS complexed with F2Y | | Descriptor: | 3,5-difluoro-L-tyrosine, Tyrosine-tRNA ligase | | Authors: | Wang, J, Tian, C, Gong, W, Li, F, Shi, P, Li, J, Ding, W. | | Deposit date: | 2012-10-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A genetically encoded 19F NMR probe for tyrosine phosphorylation.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4HJR
 
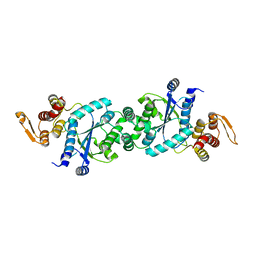 | | Crystal structure of F2YRS | | Descriptor: | Tyrosine-tRNA ligase | | Authors: | Wang, J, Tian, C, Gong, W, Li, F, Shi, P, Li, J, Ding, W. | | Deposit date: | 2012-10-13 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A genetically encoded 19F NMR probe for tyrosine phosphorylation.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7CJM
 
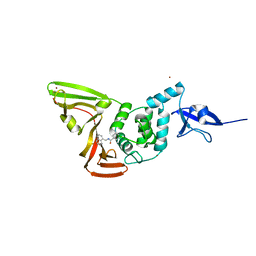 | | SARS CoV-2 PLpro in complex with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Fu, Z, Huang, H. | | Deposit date: | 2020-07-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
7UM7
 
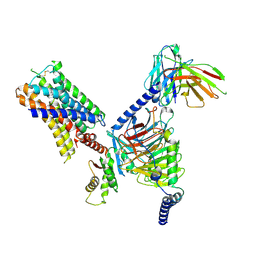 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Methylergometrine | | Descriptor: | (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM5
 
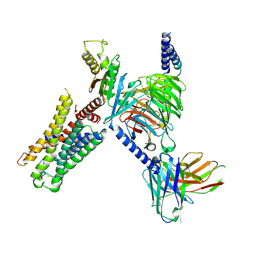 | | CryoEM structure of Go-coupled 5-HT5AR in complex with 5-CT | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM4
 
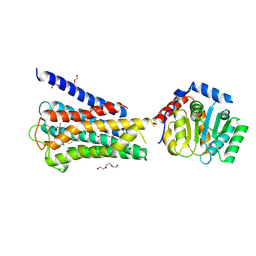 | | Crystal structure of inactive 5-HT5AR in complex with AS2674723 | | Descriptor: | 5-hydroxytryptamine receptor 5A, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Zhang, S, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UM6
 
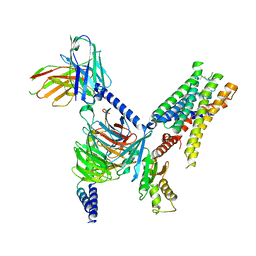 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride | | Descriptor: | 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z8B
 
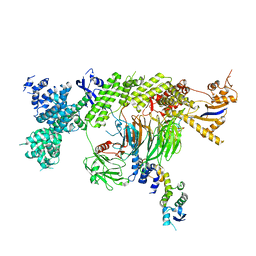 | |
5W0T
 
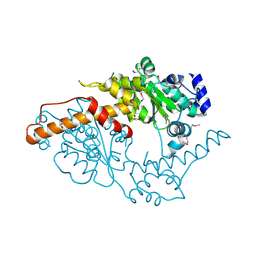 | |
