5IR6
 
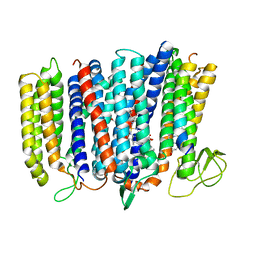 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | 分子名称: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | 著者 | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | 登録日 | 2016-03-12 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
5DOQ
 
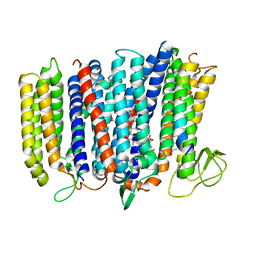 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | 分子名称: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | 著者 | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | 登録日 | 2015-09-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
7KXS
 
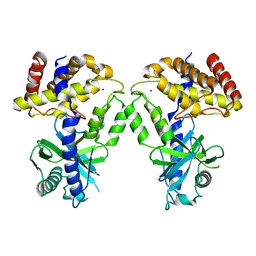 | | Computational design of constitutively active cGAS | | 分子名称: | Cyclic GMP-AMP synthase, ZINC ION | | 著者 | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | 登録日 | 2020-12-04 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7LZ3
 
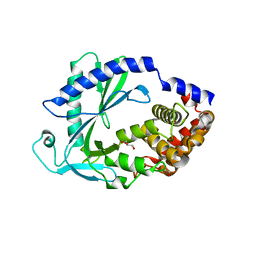 | | Computational design of constitutively active cGAS | | 分子名称: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | 著者 | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | 登録日 | 2021-03-08 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | 分子名称: | BB1 | | 著者 | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | 登録日 | 2018-04-10 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6VJY
 
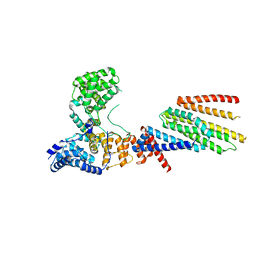 | | Cryo-EM structure of Hrd1/Hrd3 monomer | | 分子名称: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | 著者 | Wu, X, Rapoport, T.A. | | 登録日 | 2020-01-18 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
5V7V
 
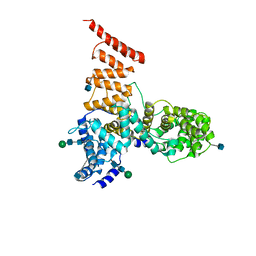 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | 登録日 | 2017-03-20 | | 公開日 | 2017-08-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
5V6P
 
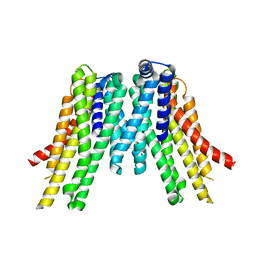 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | 分子名称: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | 著者 | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | 登録日 | 2017-03-17 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
6CZG
 
 | |
6CZJ
 
 | |
5UWB
 
 | |
9BEI
 
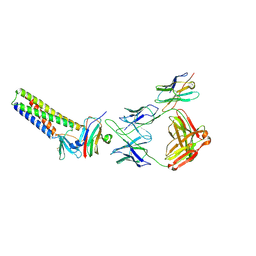 | |
8DT0
 
 | |
7AA4
 
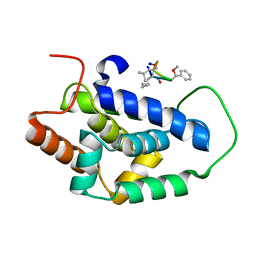 | | Structure of ClpC1-NTD bound to a CymA analogue | | 分子名称: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | 著者 | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | 登録日 | 2020-09-03 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7ABR
 
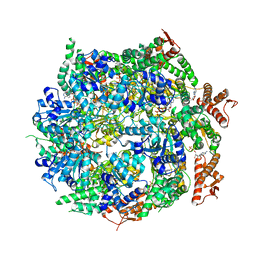 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | 著者 | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | 登録日 | 2020-09-08 | | 公開日 | 2021-10-06 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
5UVN
 
 | |
5UW2
 
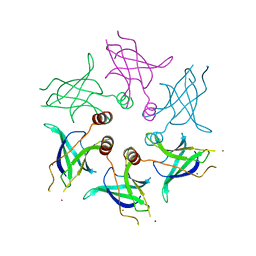 | |
5UW8
 
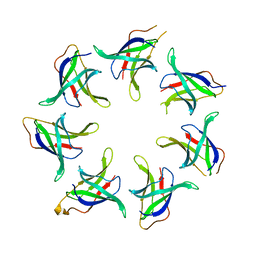 | |
7K3H
 
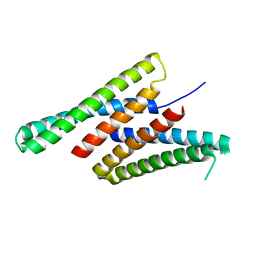 | | Crystal structure of deep network hallucinated protein 0217 | | 分子名称: | Network hallucinated protein 0217 | | 著者 | Pellock, S.J, Anishchenko, I, Chidyausiku, T.M, Bera, A.K, DiMaio, F, Baker, D. | | 登録日 | 2020-09-11 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | De novo protein design by deep network hallucination.
Nature, 600, 2021
|
|
5UWA
 
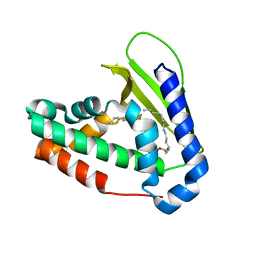 | |
8UBH
 
 | |
8SK7
 
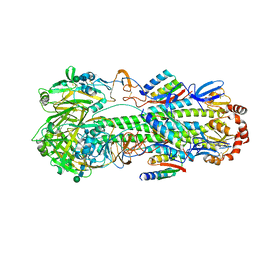 | | Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA_20 minibinder (RFdiffusion-designed), ... | | 著者 | Borst, A.J, Bennett, N.R. | | 登録日 | 2023-04-18 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | De novo design of protein structure and function with RFdiffusion.
Nature, 620, 2023
|
|
7DCN
 
 | |
7DCM
 
 | | Crystal structure of CITX | | 分子名称: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | 著者 | Xu, H, Wang, B, Su, X.D. | | 登録日 | 2020-10-26 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-05-17 | | 実験手法 | X-RAY DIFFRACTION (2.495 Å) | | 主引用文献 | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
8OYS
 
 | |
