6XAH
 
 | | Structure of a Stable Interstrand DNA Crosslink Involving an dA Amino Group and an Abasic Site | | 分子名称: | DNA (5'-D(*TP*AP*GP*AP*TP*GP*AP*AP*CP*(AAB)P*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*TP*CP*TP*AP*AP*GP*TP*TP*CP*AP*TP*CP*TP*A)-3') | | 著者 | Kellum Jr, A.H, Qiu, D, Voehler, M.W, Martin, W.J, Gates, K.S, Stone, M.P. | | 登録日 | 2020-06-04 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a Stable Interstrand DNA Cross-Link Involving a beta- N -Glycosyl Linkage Between an N 6 -dA Amino Group and an Abasic Site.
Biochemistry, 60, 2021
|
|
4X0U
 
 | |
4X0T
 
 | |
3SME
 
 | | Structure of PTP1B inactivated by H2O2/bicarbonate | | 分子名称: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Tanner, J.J, Singh, H. | | 登録日 | 2011-06-27 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Biological Buffer Bicarbonate/CO(2) Potentiates H(2)O(2)-Mediated Inactivation of Protein Tyrosine Phosphatases.
J.Am.Chem.Soc., 133, 2011
|
|
5T19
 
 | | Structure of PTP1B complexed with N-(3'-(1,1-dioxido-4-oxo-1,2,5-thiadiazolidin-2-yl)-4'-methyl-[1,1'-biphenyl]-4-yl)acetamide | | 分子名称: | 5-[4-methyl-4'-(methylamino)[1,1'-biphenyl]-3-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Laciak, A.R, Tanner, J.J. | | 登録日 | 2016-08-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1001 Å) | | 主引用文献 | Covalent Allosteric Inactivation of Protein Tyrosine Phosphatase 1B (PTP1B) by an Inhibitor-Electrophile Conjugate.
Biochemistry, 56, 2017
|
|
6U2X
 
 | |
6UFP
 
 | | Structure of proline utilization A with the FAD covalently modified by L-thiazolidine-2-carboxylate and three cysteines (Cys46, Cys470, Cys638) modified to S,S-(2-HYDROXYETHYL)THIOCYSTEINE | | 分子名称: | (2S)-1,3-thiazolidine-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Campbell, A.C, Tanner, J.J. | | 登録日 | 2019-09-24 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.737 Å) | | 主引用文献 | Covalent Modification of the Flavin in Proline Dehydrogenase by Thiazolidine-2-Carboxylate.
Acs Chem.Biol., 15, 2020
|
|
6VR6
 
 | |
6VZ9
 
 | |
6VWF
 
 | |
7MY9
 
 | | Structure of proline utilization A with 1,3-dithiolane-2-carboxylate bound in the proline dehydrogenase active site | | 分子名称: | 1,3-dithiolane-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tanner, J.J, Campbell, A.C. | | 登録日 | 2021-05-20 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.628 Å) | | 主引用文献 | Photoinduced Covalent Irreversible Inactivation of Proline Dehydrogenase by S-Heterocycles.
Acs Chem.Biol., 16, 2021
|
|
7MYB
 
 | |
7MYA
 
 | |
7MYC
 
 | |
6O4E
 
 | | Structure of ALDH7A1 mutant N167S complexed with NAD | | 分子名称: | Alpha-aminoadipic semialdehyde dehydrogenase, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4F
 
 | | Structure of ALDH7A1 mutant N167S complexed with alpha-aminoadipate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4H
 
 | | Structure of ALDH7A1 mutant A171V complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4G
 
 | |
6O4C
 
 | | Structure of ALDH7A1 mutant W175A complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4K
 
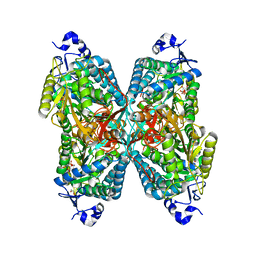 | | Structure of ALDH7A1 mutant E399Q complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4I
 
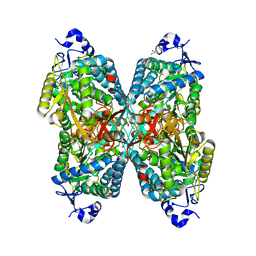 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4D
 
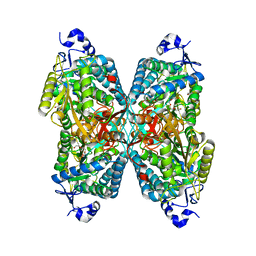 | | Structure of ALDH7A1 mutant W175A complexed with L-pipecolic acid | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4B
 
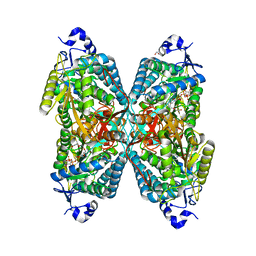 | | Structure of ALDH7A1 mutant W175G complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4L
 
 | | Structure of ALDH7A1 mutant E399D complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
7JL5
 
 | | Crystal structure of human NEIL3 tandem zinc finger GRF domains | | 分子名称: | Endonuclease 8-like 3, ZINC ION | | 著者 | Rodriguez, A.A, Wojtaszek, J.L, Williams, R.S, Eichman, B.F. | | 登録日 | 2020-07-29 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An autoinhibitory role for the GRF zinc finger domain of DNA glycosylase NEIL3.
J.Biol.Chem., 295, 2020
|
|
