6QCM
 
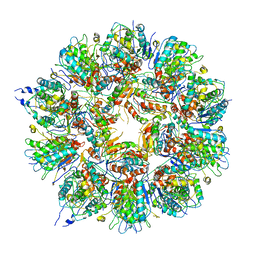 | | Cryo em structure of the Listeria stressosome | | Descriptor: | RsbR protein, RsbR protein,RsbR protein, RsbS protein | | Authors: | Williams, A.H, Redzej, A, Waksman, G, Cossart, P. | | Deposit date: | 2018-12-28 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation.
Nat Commun, 10, 2019
|
|
4WAV
 
 | | Crystal Structure of Haloquadratum walsbyi bacteriorhodopsin mutant D93N | | Descriptor: | Bacteriorhodopsin-I, RETINAL, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Wang, A.H.J, Hsu, M.F, Yang, C.S, Fu, H.Y. | | Deposit date: | 2014-09-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an acid-tolerant light-driven proton pump at 1.85 Angstroms resolution
To be published
|
|
4OBV
 
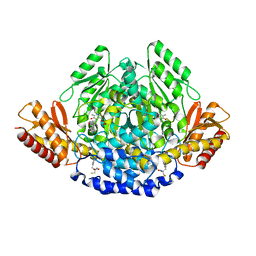 | | Ruminococcus gnavus tryptophan decarboxylase RUMGNA_01526 (alpha-FMT) | | Descriptor: | Pyridoxal-dependent decarboxylase domain protein, alpha-(fluoromethyl)-D-tryptophan, {5-hydroxy-4-[(1E)-4-(1H-indol-3-yl)-3-oxobut-1-en-1-yl]-6-methylpyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Fraser, J.S, Van Benschoten, A.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine.
Cell Host Microbe, 16, 2014
|
|
4V5N
 
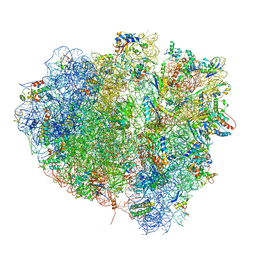 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
5DBJ
 
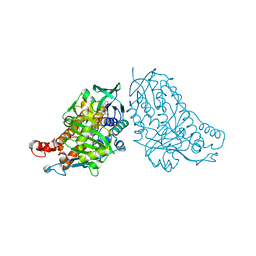 | | Crystal structure of halogenase PltA | | Descriptor: | CHLORIDE ION, FADH2-dependent halogenase PltA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pang, A.H, Tsodikov, O.V. | | Deposit date: | 2015-08-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of halogenase PltA from the pyoluteorin biosynthetic pathway.
J.Struct.Biol., 192, 2015
|
|
8EBZ
 
 | |
2YM7
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 5-({6-[(piperidin-4-ylmethyl)amino]pyrimidin-4-yl}amino)pyrazine-2-carbonitrile, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
6SXI
 
 | | Antibody-anti-idiotype complex: AP33 Fab (hepatitis C virus E2 antibody) - B2.1A scFv (anti-idiotype) | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Taylor, G.L, Potter, J.A, Fadda, V, Patel, A.H, Owsianka, A.M, Cowtan, V.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-10-07 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of a structural epitope mimic: an idiotypic approach to HCV vaccine design.
NPJ Vaccines, 6, 2021
|
|
7NRP
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3'), RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-03-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
6DCS
 
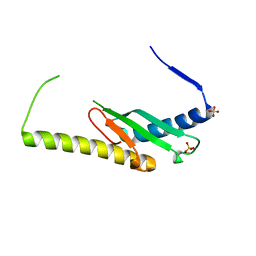 | | Stage III sporulation protein AF (SpoIIIAF) | | Descriptor: | SULFATE ION, Stage III sporulation protein AF | | Authors: | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | Deposit date: | 2018-05-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6DV5
 
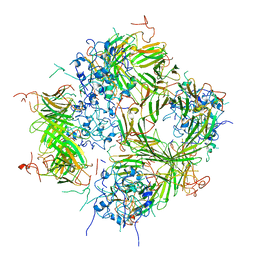 | |
8SZO
 
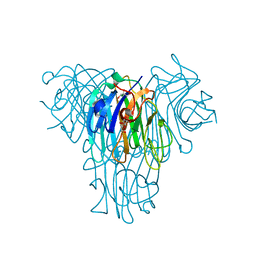 | | Canavalia villosa lectin in complex with alpha-methyl-mannoside | | Descriptor: | CALCIUM ION, Canavalia villosa lectin, GLYCEROL, ... | | Authors: | Cavada, B.S, Lossio, C.F, Pinto-Junior, V.R, Osterne, V.J.S, Oliveira, M.V, Neco, A.H.B, Nascimento, K.S. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lectin from Canavalia villosa seeds: A glucose/mannose-specific protein and a new tool for inflammation studies.
Int J Biol Macromol, 105, 2017
|
|
3UHA
 
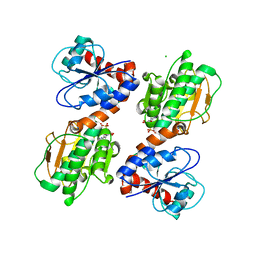 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cervisiae complexed with NAD. | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Saccharopine dehydrogenase [NAD+, ... | | Authors: | Cook, P.F, Kumar, V.P, Thomas, L.M, West, A.H, Bobyk, K.D. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
5KDE
 
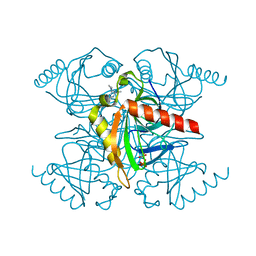 | | Inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with inhibitor 1 and inorganic pyrophosphate | | Descriptor: | 2,4-bis(aziridin-1-yl)-6-(1-phenylpyrrol-2-yl)-1,3,5-triazine, Inorganic pyrophosphatase, PYROPHOSPHATE 2- | | Authors: | Pang, A.H, Garzan, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of Allosteric and Selective Inhibitors of Inorganic Pyrophosphatase from Mycobacterium tuberculosis.
ACS Chem. Biol., 11, 2016
|
|
3UH1
 
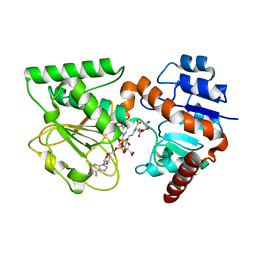 | | Crystal Structure of Saccharopine Dehydrogenase from Saccharomyces cerevisiae with bound saccharopine and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, ... | | Authors: | Kumar, V.P, Thomas, L.M, Bobyk, K.D, Andi, B, West, A.H, Cook, P.F. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
8STR
 
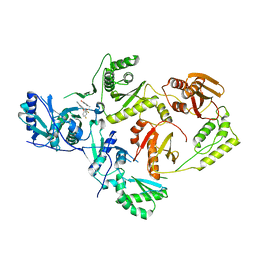 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Chan, A.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
3UGK
 
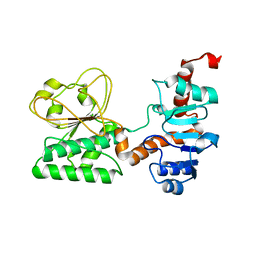 | | Crystal Structure of C205S mutant and Saccharopine Dehydrogenase from Saccharomyces cerevisiae. | | Descriptor: | Saccharopine dehydrogenase [NAD+, L-lysine-forming] | | Authors: | Cook, P.F, Kumar, V.P, Thomas, L.M, West, A.H, Bobyk, K.D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Evidence in Support of Lysine 77 and Histidine 96 as Acid-Base Catalytic Residues in Saccharopine Dehydrogenase from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
5KDF
 
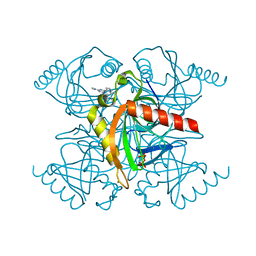 | | Inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with inhibitor 6 and inorganic pyrophosphate | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, PYROPHOSPHATE 2-, ... | | Authors: | Pang, A.H, Garzan, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Allosteric and Selective Inhibitors of Inorganic Pyrophosphatase from Mycobacterium tuberculosis.
ACS Chem. Biol., 11, 2016
|
|
5K6P
 
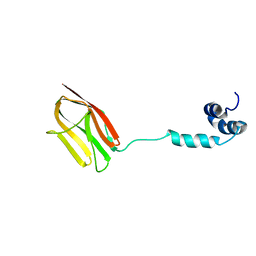 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
8TW0
 
 | | Crystal Structure of a synthetic ABC heterotrimeric Collagen-like Peptide at 1.53 A | | Descriptor: | Collagen Mimetic Peptide A, Collagen Mimetic Peptide B, Collagen Mimetic Peptide C, ... | | Authors: | Miller, M.D, Cole, C.C, Xu, W, Walker, D.R, Hulgan, S.A.H, Pogostin, B.H, Swain, J.W.R, Duella, R, Misiura, M, Wang, X, Kolomeisky, A.B, Phillips Jr, G.N, Hartgerink, J.D. | | Deposit date: | 2023-08-18 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Heterotrimeric Collagen Helix with High Specificity of Assembly Results in a Rapid Rate of Folding
Nat.Chem., 2024
|
|
7RGW
 
 | |
7OOO
 
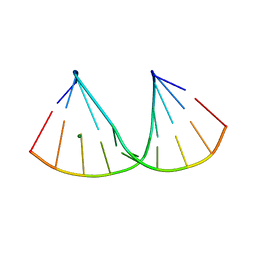 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG containing an LNA-Amide-LNA modification | | Descriptor: | DNA (5'-D(*CP*TP*(05A)P*TP*CP*TP*TP*TP*G)-3'), MAGNESIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OOS
 
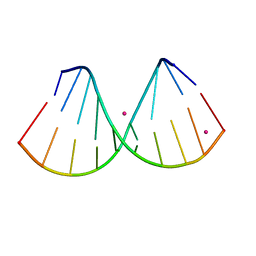 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG | | Descriptor: | DNA (5'-D(*CP*TP*(05K)P*TP*CP*TP*TP*TP*G)-3'), RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), STRONTIUM ION | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OZZ
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG with LNA-amide modification | | Descriptor: | DNA (5'-D(*CP*TP*(05H)P*TP*CP*TP*TP*TP*G)-3'), POTASSIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-06-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7SO6
 
 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
