4PP4
 
 | |
6KBE
 
 | | Structure of Deubiquitinase | | 分子名称: | Polyubiquitin-C, Ubiquitin thioesterase | | 著者 | Lu, L.N, Liu, L, Wang, F. | | 登録日 | 2019-06-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.339 Å) | | 主引用文献 | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6K9P
 
 | | Structure of Deubiquitinase | | 分子名称: | Ubiquitin, Ubiquitin thioesterase | | 著者 | Lu, L.N, Liu, L, Wang, F. | | 登録日 | 2019-06-17 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.047 Å) | | 主引用文献 | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
8HR1
 
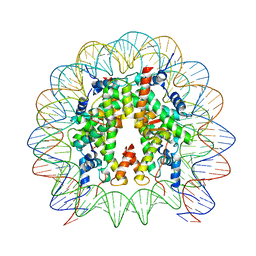 | | Cryo-EM structure of SSX1 bound to the unmodified nucleosome at a resolution of 3.02 angstrom | | 分子名称: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Zebin, T, Ai, H.S, Ziyu, X, Man, P, Liu, L. | | 登録日 | 2022-12-14 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQY
 
 | | Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom | | 分子名称: | DNA (136-MER), DNA (137-MER), Histone H2A type 1-B/E, ... | | 著者 | Zebin, T, Ai, H.S, Ziyu, X, GuoChao, C, Man, P, Liu, L. | | 登録日 | 2022-12-14 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GMT
 
 | | Structure of UmuD in complex with RecA filament | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA polymerase V subunit UmuD, MAGNESIUM ION, ... | | 著者 | Gao, B, Feng, Y. | | 登録日 | 2022-08-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GMU
 
 | |
8GMS
 
 | | Structure of LexA in complex with RecA filament | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), LexA repressor, MAGNESIUM ION, ... | | 著者 | Gao, B, Feng, Y. | | 登録日 | 2022-08-22 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ISU
 
 | | Crystal structure of Lys27-linked di-ubiquitin in complex with its selective interacting protein UCHL3 | | 分子名称: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | 著者 | Ding, S, Pan, M, Zheng, Q, Ren, Y, Hong, D. | | 登録日 | 2018-11-19 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.866 Å) | | 主引用文献 | Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6K6L
 
 | | YGL082W-catalytic domain | | 分子名称: | pseudo deubiquitinase | | 著者 | Lu, L.N, Wang, F. | | 登録日 | 2019-06-03 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Inactivity of YGL082W in vitro due to impairment of conformational change in the catalytic center loop
Sci China Chem, 2019
|
|
5C5P
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (3R)-3-(1-hydroxy-2-methylpropan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5R
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (7R)-2-hydroxy-7-(propan-2-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine-3-carbonitrile, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5Q
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (3R)-10-methyl-3-(propan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
8UCD
 
 | | Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, AMG 509 anti-STEAP1 Fab, heavy chain, ... | | 著者 | Li, F, Bailis, J.M, Zhang, H. | | 登録日 | 2023-09-26 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | AMG 509 (Xaluritamig), an Anti-STEAP1 XmAb 2+1 T-cell Redirecting Immune Therapy with Avidity-Dependent Activity against Prostate Cancer.
Cancer Discov, 14, 2024
|
|
3G6L
 
 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | 分子名称: | CAFFEINE, Chitinase | | 著者 | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | 登録日 | 2009-02-06 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
5XDK
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with CO-1686 | | 分子名称: | Epidermal growth factor receptor, N-[3-[[2-[[4-(4-ethanoylpiperazin-1-yl)-2-methoxy-phenyl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide | | 著者 | Yan, X.E, Yun, C.H. | | 登録日 | 2017-03-28 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.346 Å) | | 主引用文献 | Structural basis of mutant-selectivity and drug-resistance related to CO-1686.
Oncotarget, 8, 2017
|
|
5XDL
 
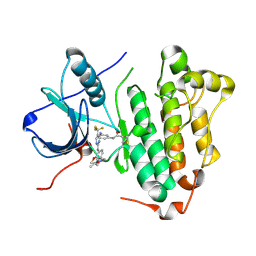 | |
8K8T
 
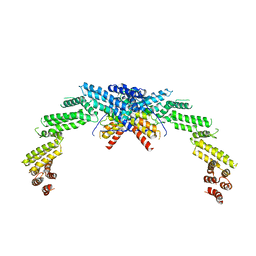 | | Structure of CUL3-RBX1-KLHL22 complex | | 分子名称: | Cullin-3, Kelch-like protein 22 | | 著者 | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | 登録日 | 2023-07-31 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | 著者 | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | 登録日 | 2023-08-01 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
4TTH
 
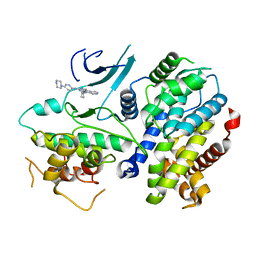 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | 分子名称: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | 著者 | Piper, D.E, Walker, N, Wang, Z. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-25 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
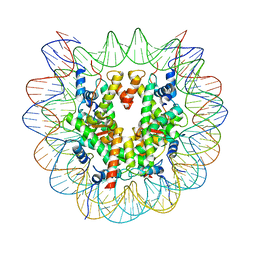 | | cryo-EM structure of unmodified nucleosome | | 分子名称: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-26 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XCR
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-25 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD0
 
 | | cryo-EM structure of H2BK34ub nucleosome | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-26 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
