8EPV
 
 | | 2.2 A crystal structure of the lipocalin cat allergen Fel d 7 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Fel d 7 allergen, ... | | 著者 | Min, J, Pedersen, L.C, Geoffrey, M.A. | | 登録日 | 2022-10-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural and ligand binding analysis of the pet allergens Can f 1 and Fel d 7.
Front Allergy, 4, 2023
|
|
8U0O
 
 | | Synaptic complex of human DNA polymerase Lambda DL variant engaged on a DNA double-strand break containing an unpaired 3' primer terminus | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*CP*GP*CP*GP*GP*CP*A)-3'), ... | | 著者 | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Kunkel, T.A, Chiruvella, K.K, Ramsden, D.A. | | 登録日 | 2023-08-29 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | DNA polymerase lambda Loop1 variant yields unexpected gain-of-function capabilities in nonhomologous end-joining.
DNA Repair (Amst), 136, 2024
|
|
8U0P
 
 | | Synaptic complex of human DNA polymerase Lambda DL variant engaged on a noncomplementary DNA double-strand break | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | 著者 | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Kunkel, T.A, Chiruvella, K.K, Ramsden, D.A. | | 登録日 | 2023-08-29 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | DNA polymerase lambda Loop1 variant yields unexpected gain-of-function capabilities in nonhomologous end-joining.
DNA Repair (Amst), 136, 2024
|
|
2FJ6
 
 | | Solution NMR structure of the UPF0346 protein yozE from Bacillus subtilis. Northeast Structural Genomics target SR391. | | 分子名称: | Hypothetical UPF0346 protein yozE | | 著者 | Rossi, P, Acton, T.B, Cunningham, K.E, Ma, L.C, Shetty, K, Swapna, G.V.T, Xiao, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-12-31 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the UPF0346 protein yozE from Bacillus subtilis. Northeast Structural Genomics target SR391.
To be Published
|
|
2EST
 
 | |
2FMS
 
 | | DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | 著者 | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | 登録日 | 2006-01-09 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FMQ
 
 | | Sodium in active site of DNA Polymerase Beta | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | 著者 | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | 登録日 | 2006-01-09 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FVT
 
 | | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris: Northeast Structural Genomics Target RpR43 | | 分子名称: | conserved hypothetical protein | | 著者 | Cort, J.R, Ho, C.K, Cunningham, K, Ma, L.C, Conover, K, Xiao, R, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-01-31 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris
To be Published
|
|
2FMP
 
 | | DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | 著者 | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | 登録日 | 2006-01-09 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
3H0O
 
 | | The importance of CH-Pi stacking interactions between carbohydrate and aromatic residues in truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | 著者 | Tsai, L.C, Hsiao, C.H. | | 登録日 | 2009-04-09 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The importance of CH-Pi stacking interactions between carbohydrate and aromatic residues in truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
To be Published
|
|
3HR9
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase F40I mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | 著者 | Tsai, L.C, Huang, H.C, Hsiao, C.H. | | 登録日 | 2009-06-09 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase mutant F40I
To be Published
|
|
1TW7
 
 | | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target | | 分子名称: | SODIUM ION, protease | | 著者 | Martin, P, Vickrey, J.F, Proteasa, G, Jimenez, Y.L, Wawrzak, Z, Winters, M.A, Merigan, T.C, Kovari, L.C. | | 登録日 | 2004-06-30 | | 公開日 | 2005-07-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Wide Open 1.3A Structure of a Multi-drug Resistant HIV-1 Protease Represents a Novel Drug Target
Structure, 13, 2005
|
|
8OWA
 
 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-4 | | 分子名称: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | 著者 | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-21 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
8OWL
 
 | | SR Ca(2+)-ATPase in the E2 state complexed with the photoswitch-thapsigargin derivative AzTG-6 | | 分子名称: | ACETYL GROUP, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1, ... | | 著者 | Hjorth-Jensen, S.J, Konrad, D.B, Quistgaard, E.M.H, Hansen, L.C, Novak, A, Chu, H, Jurasek, M, Zimmermann, T, Andersen, J.L, Baran, P.S, Nissen, P, Trauner, D. | | 登録日 | 2023-04-28 | | 公開日 | 2023-06-21 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Photoswitchable inhibitors of the sarco(endo)plasmic calcium pump
To Be Published
|
|
7BH8
 
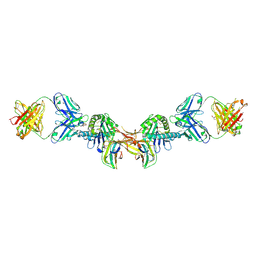 | | 3H4-Fab HLA-E-VL9 co-complex | | 分子名称: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | 著者 | Walters, L.C, Rozbesky, D. | | 登録日 | 2021-01-10 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
4M04
 
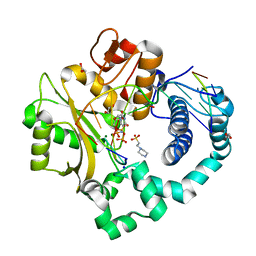 | | Human DNA Polymerase Mu ternary complex | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | 登録日 | 2013-08-01 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4LZD
 
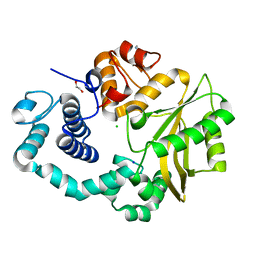 | | Human DNA polymerase mu- Apoenzyme | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, ... | | 著者 | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | 登録日 | 2013-07-31 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4LZG
 
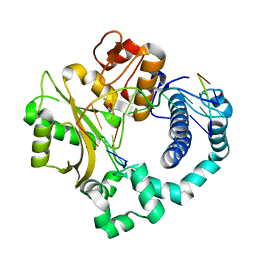 | | Binary complex of human DNA Polymerase Mu with DNA | | 分子名称: | CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, GLYCEROL, ... | | 著者 | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | 登録日 | 2013-07-31 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7EHJ
 
 | | human MTHFD2 in complex with compound 21, cofactor and phosphate. | | 分子名称: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | 著者 | Lee, L.C, Peng, Y.H, Wu, S.Y. | | 登録日 | 2021-03-29 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHM
 
 | | Human MTHFD2 in complex with compound 21 and 15 | | 分子名称: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, (2S)-2-[[4-[[1-[(3,4-dichlorophenyl)methyl]-3,7-dimethyl-2,6-bis(oxidanylidene)purin-8-yl]amino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | 著者 | Lee, L.C, Peng, Y.H, Wu, S.Y. | | 登録日 | 2021-03-30 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHN
 
 | | Human MTHFD2 in complex with compound 21 and 9 | | 分子名称: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 3-[4-[[1-[(4-chloranyl-1H-indol-2-yl)methyl]-3,7-dimethyl-2,6-bis(oxidanylidene)purin-8-yl]amino]-6-methyl-pyrimidin-2-yl]propanoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | 著者 | Lee, L.C, Peng, Y.H, Wu, S.Y. | | 登録日 | 2021-03-30 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
7EHV
 
 | | Human MTHFD2 in complex with compound 21 and 3 | | 分子名称: | (2S)-2-[[4-[(4-azanyl-6-oxidanyl-pyrimidin-5-yl)carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 1-(3,4-dichlorobenzyl)-8-(((1R,4R)-4-hydroxycyclohexyl)amino)-3,7-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | 著者 | Lee, L.C, Peng, Y.H, Wu, S.Y. | | 登録日 | 2021-03-30 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Xanthine Derivatives Reveal an Allosteric Binding Site in Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2).
J.Med.Chem., 64, 2021
|
|
5Z2G
 
 | | Crystal Structure of L-amino acid oxidase from venom of Naja atra | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase | | 著者 | Kumar, J.V, Chien, K.Y, Wu, W.G, Lin, C.C, Chiang, L.C, Lin, T.H. | | 登録日 | 2018-01-02 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.676 Å) | | 主引用文献 | Crystal Structure of L-amino acid oxidase from naja atra (Taiwan Cobra)
To Be Published
|
|
5W7X
 
 | |
6A6O
 
 | | Crystal structure of acetyl ester-xyloside bifunctional hydrolase from Caldicellulosiruptor lactoaceticus | | 分子名称: | Esterase/lipase-like protein | | 著者 | Cao, H, Huang, Y, Sun, L.C, Liu, X, Liu, T.F, Wang, F.Z, Xin, F.J. | | 登録日 | 2018-06-28 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Insights into the Dual-Substrate Recognition and Catalytic Mechanisms of a Bifunctional Acetyl Ester-Xyloside Hydrolase from Caldicellulosiruptor lactoaceticus.
Acs Catalysis, 9, 2019
|
|
