4NZZ
 
 | |
7CMI
 
 | | The LAT2-4F2hc complex in complex with leucine | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | 著者 | Yan, R.H, Zhou, J.Y, Li, Y.N, Lei, J.L, Zhou, Q. | | 登録日 | 2020-07-27 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex.
Cell Discov, 6, 2020
|
|
1Y4K
 
 | | Lipoxygenase-1 (Soybean) at 100K, N694G Mutant | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | 著者 | Chruszcz, M, Segraves, E, Holman, T.R, Minor, W. | | 登録日 | 2004-12-01 | | 公開日 | 2005-12-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly.
Biochemistry, 45, 2006
|
|
8JAN
 
 | |
8JAJ
 
 | |
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | 著者 | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | 登録日 | 2023-05-13 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
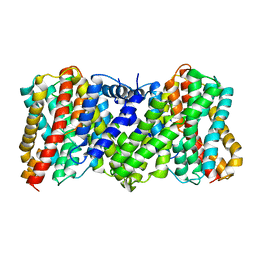 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | 分子名称: | Sodium/hydrogen exchanger 7 | | 著者 | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | 登録日 | 2023-05-13 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
5HSJ
 
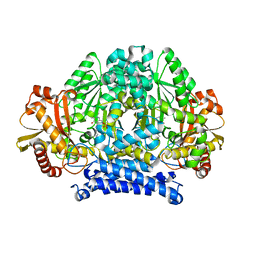 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | 著者 | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | 登録日 | 2016-01-25 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
5XFS
 
 | |
6MBE
 
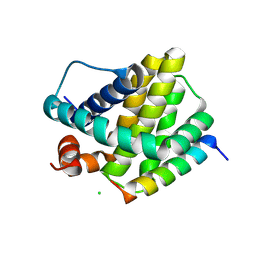 | | Human Mcl-1 in complex with the designed peptide dM7 | | 分子名称: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | 著者 | Jenson, J.M, Keating, A.E. | | 登録日 | 2018-08-29 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
8JVA
 
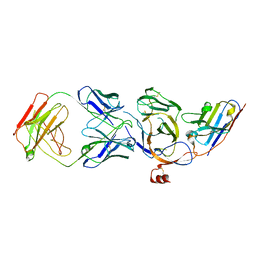 | | Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 heavy chain, S2L20 light chain, ... | | 著者 | Liu, B, Liu, H.H, Han, P, Qi, J.X. | | 登録日 | 2023-06-28 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Enhanced potency of an IgM-like nanobody targeting conserved epitope in SARS-CoV-2 spike N-terminal domain.
Signal Transduct Target Ther, 9, 2024
|
|
7X0B
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2022-02-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02027535 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZX
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2022-02-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.980013 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | 分子名称: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | 著者 | Zhou, J.H, Hou, X.L. | | 登録日 | 2022-02-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.899313 Å) | | 主引用文献 | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
5Z0Q
 
 | | Crystal Structure of OvoB | | 分子名称: | Aminotransferase, class I and II, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Cai, Y.J, Huang, P, Wu, L, Zhou, J.H, Liu, P.H. | | 登録日 | 2017-12-20 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | In Vitro Reconstitution of the Remaining Steps in Ovothiol A Biosynthesis: C-S Lyase and Methyltransferase Reactions.
Org. Lett., 20, 2018
|
|
5ZOL
 
 | | Crystal structure of a three sites mutantion of FSAA complexed with HA and product | | 分子名称: | (3S,4S)-3,4-dihydroxy-4-(thiophen-2-yl)butan-2-one, 1-hydroxypropan-2-one, CHLORIDE ION, ... | | 著者 | Wu, L, Yang, X.H, Yu, H.W, Zhou, J.H. | | 登録日 | 2018-04-13 | | 公開日 | 2019-06-12 | | 最終更新日 | 2020-07-22 | | 実験手法 | X-RAY DIFFRACTION (2.172 Å) | | 主引用文献 | The engineering of decameric d-fructose-6-phosphate aldolase A by combinatorial modulation of inter- and intra-subunit interactions.
Chem.Commun.(Camb.), 56, 2020
|
|
7VOC
 
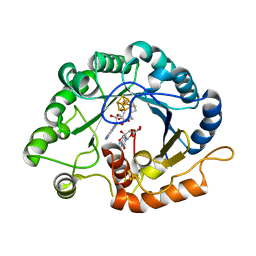 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | 分子名称: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | 著者 | Hou, X.L, Zhou, J.H. | | 登録日 | 2021-10-13 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.62005424 Å) | | 主引用文献 | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
7VOB
 
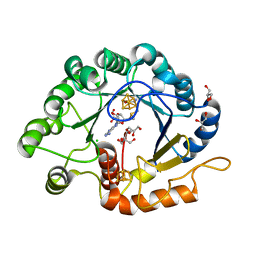 | |
7W5N
 
 | | The crystal structure of the reduced form of Gluconobacter oxydans WSH-004 SNDH | | 分子名称: | L-sorbosone dehydrogenase, NAD(P) dependent, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, D, Hou, X.D, Rao, Y.J, Yin, D.J, Zhou, J.W, Chen, J. | | 登録日 | 2021-11-30 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.988 Å) | | 主引用文献 | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7W5L
 
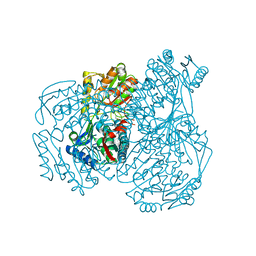 | | The crystal structure of the oxidized form of Gluconobacter oxydans WSH-004 SNDH | | 分子名称: | L-sorbosone dehydrogenase, NAD(P) dependent | | 著者 | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | 登録日 | 2021-11-30 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7DL3
 
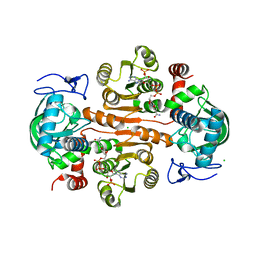 | | The structure of 3,5-DAHDHcca complex with NADPH | | 分子名称: | 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-25 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84606934 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL0
 
 | | The mutant E310G/A314Y of 3,5-DAHDHcca complex with NADPH | | 分子名称: | 1,2-ETHANEDIOL, 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, ... | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-25 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL7
 
 | | The wild-type structure of 3,5-DAHDHcca | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-diaminohexanoate dehydrogenase, ... | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-26 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.30065823 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL1
 
 | | The mutant E310G/G323S structure of 3,5-DAHDHcca complex with NADPH | | 分子名称: | 3,5-diaminohexanoate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | 登録日 | 2020-11-25 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6JJJ
 
 | |
