3PBQ
 
 | | Crystal structure of PBP3 complexed with imipenem | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | 著者 | Han, S. | | 登録日 | 2010-10-20 | | 公開日 | 2010-12-22 | | 最終更新日 | 2011-08-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4ZZA
 
 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with raffinose, selenomethionine derivative | | 分子名称: | Sugar binding protein of ABC transporter system, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | 著者 | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Abou Hachem, M. | | 登録日 | 2015-05-22 | | 公開日 | 2016-06-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
3PD9
 
 | | X-ray structure of the ligand-binding core of GluA2 in complex with (R)-5-HPCA at 2.1 A resolution | | 分子名称: | (5R)-3-hydroxy-4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Frydenvang, K, Kastrup, J.S. | | 登録日 | 2010-10-22 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Biostructural and pharmacological studies of bicyclic analogues of the 3-isoxazolol glutamate receptor agonist ibotenic acid.
J. Med. Chem., 53, 2010
|
|
3PBR
 
 | | Crystal structure of PBP3 complexed with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | 著者 | Han, S. | | 登録日 | 2010-10-20 | | 公開日 | 2010-12-22 | | 最終更新日 | 2012-05-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MWV
 
 | | Crystal structure of HCV NS5B polymerase | | 分子名称: | Genome polyprotein | | 著者 | Coulombe, R. | | 登録日 | 2010-05-06 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
3MXO
 
 | | Crystal structure oh human phosphoglycerate mutase family member 5 (PGAM5) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-05-07 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
4F08
 
 | |
3MWW
 
 | | Crystal structure of HCV NS5B polymerase | | 分子名称: | 1-[2-(4-carboxypiperidin-1-yl)-2-oxoethyl]-3-cyclohexyl-2-furan-3-yl-1H-indole-6-carboxylic acid, Genome polyprotein, SULFATE ION | | 著者 | Coulombe, R. | | 登録日 | 2010-05-06 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
3MX6
 
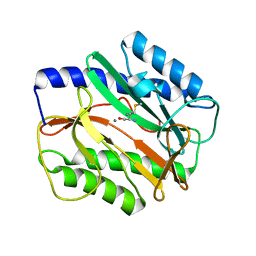 | |
3MYG
 
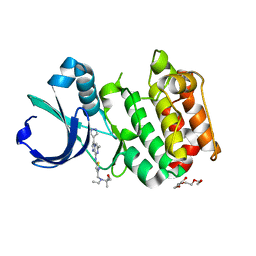 | | Aurora A Kinase complexed with SCH 1473759 | | 分子名称: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | 著者 | Hruza, A, Prosis, W, Ramanathan, L. | | 登録日 | 2010-05-10 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
5CUS
 
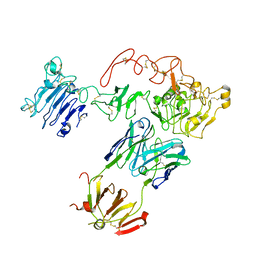 | | Crystal Structure of sErbB3-Fab3379 Complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab LC region of KTN3379, IgG H chain, ... | | 著者 | Lee, S, Schlessinger, J. | | 登録日 | 2015-07-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5D6K
 
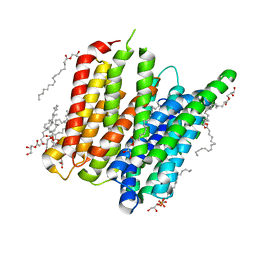 | | PepT - CIM | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, Di-or tripeptide:H+ symporter, ... | | 著者 | Ma, P, Caffrey, M. | | 登録日 | 2015-08-12 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
4CAV
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a benzofuran ligand R0-09-4879 | | 分子名称: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, CHLORIDE ION, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | 著者 | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | 登録日 | 2013-10-09 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
5D6L
 
 | | beta2AR-T4L - CIM | | 分子名称: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | 著者 | Ma, P, Caffrey, M. | | 登録日 | 2015-08-12 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
3OII
 
 | |
3OIJ
 
 | |
3OI7
 
 | | Structure of the structure of the H13A mutant of Ykr043C in complex with sedoheptulose-1,7-bisphosphate | | 分子名称: | 1,2-ETHANEDIOL, 1,7-di-O-phosphono-beta-D-altro-hept-2-ulofuranose, GLYCEROL, ... | | 著者 | Singer, A.U, Xu, X, Dong, A, Cui, H, Clasquin, M.F, Caudy, A.A, Edwards, A.M, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-08-18 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Riboneogenesis in yeast.
Cell(Cambridge,Mass.), 145, 2011
|
|
3OIN
 
 | |
5CXT
 
 | |
5D1R
 
 | | Crystal structure of Mycobacterium tuberculosis Rv1816 transcriptional regulator. | | 分子名称: | MAGNESIUM ION, NICKEL (II) ION, Rv1816 transcriptional regulator, ... | | 著者 | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | 登録日 | 2015-08-04 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4DVW
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with MAE-II-167 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-chloro-3-fluorophenyl)-N'-[(3aS,6aS)-hexahydrocyclopenta[c]pyrrol-3a(1H)-ylmethyl]ethanediamide, ... | | 著者 | Kwon, Y.D, Kwong, P.D. | | 登録日 | 2012-02-23 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
4DA5
 
 | | Choline Kinase alpha acts through a double-displacement kinetic mechanism involving enzyme isomerisation, as determined through enzyme and inhibitor kinetics and structural biology | | 分子名称: | (3R)-1-azabicyclo[2.2.2]oct-3-yl[bis(5-chlorothiophen-2-yl)]methanol, Choline kinase alpha, SULFATE ION | | 著者 | Brown, K, Hudson, C, Charlton, P, Pollard, J. | | 登録日 | 2012-01-12 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Kinetic and mechanistic characterisation of Choline Kinase-alpha.
Biochim.Biophys.Acta, 1834, 2013
|
|
4DCE
 
 | | Crystal structure of human anaplastic lymphoma kinase in complex with a piperidine-carboxamide inhibitor | | 分子名称: | (3S)-N-(4-methylbenzyl)-1-{2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}piperidine-3-carboxamide, ALK tyrosine kinase receptor | | 著者 | Whittington, D.A, Epstein, L.F, Chen, H. | | 登録日 | 2012-01-17 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Rapid development of piperidine carboxamides as potent and selective anaplastic lymphoma kinase inhibitors.
J.Med.Chem., 55, 2012
|
|
5AQ5
 
 | |
4DMN
 
 | | HIV-1 Integrase Catalytical Core Domain | | 分子名称: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](methoxy)ethanoic acid, ARSENIC, HIV-1 Integrase, ... | | 著者 | Feng, L, Kvaratskhelia, M. | | 登録日 | 2012-02-08 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors.
J.Biol.Chem., 287, 2012
|
|
