1FWX
 
 | | CRYSTAL STRUCTURE OF NITROUS OXIDE REDUCTASE FROM P. DENITRIFICANS | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Brown, K, Djinovic-Carugo, K, Haltia, T, Cabrito, I, Saraste, M, Moura, J.J, Moura, I, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Revisiting the Catalytic CuZ Cluster of Nitrous Oxide (N2O) Reductase. Evidence of a Bridging Inorganic Sulfur
J.Biol.Chem., 275, 2000
|
|
1TYB
 
 | |
1TYA
 
 | |
1TYC
 
 | |
2KH7
 
 | |
2KH8
 
 | |
5HJ2
 
 | | Integrin alpha2beta1 I-domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brown, K.L, Banerjee, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Salt-bridge modulates differential calcium-mediated ligand binding to integrin alpha 1- and alpha 2-I domains.
Sci Rep, 8, 2018
|
|
1HZU
 
 | | DOMAIN SWING UPON HIS TO ALA MUTATION IN NITRITE REDUCTASE OF PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRITE REDUCTASE | | Authors: | Brown, K, Cutruzzola, F, Brunori, M, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-01-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.
J.Mol.Biol., 312, 2001
|
|
1HZV
 
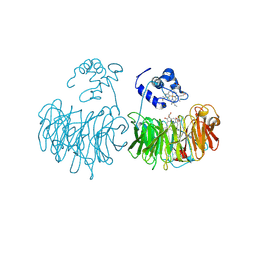 | | DOMAIN SWING UPON HIS TO ALA MUTATION IN NITRITE REDUCTASE OF PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-01-26 | | Release date: | 2001-09-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.
J.Mol.Biol., 312, 2001
|
|
5HGJ
 
 | | Structure of integrin alpha1beta1 and alpha2beta1 I-domains explain differential calcium-mediated ligand recognition | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Brown, K.L, Banerjee, S, Feigley, A, Abe, H, Blackwell, T, Zent, R, Pozzi, A, Hudson, B.H. | | Deposit date: | 2016-01-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Salt-bridge modulates differential calcium-mediated ligand binding to integrin alpha 1- and alpha 2-I domains.
Sci Rep, 8, 2018
|
|
1CNO
 
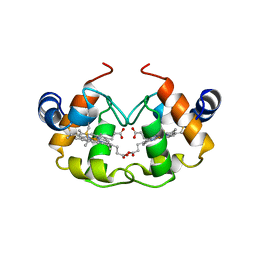 | | STRUCTURE OF PSEUDOMONAS NAUTICA CYTOCHROME C552, BY MAD METHOD | | Descriptor: | CYTOCHROME C552, GLYCEROL, HEME C | | Authors: | Brown, K, Nurizzo, D, Cambillau, C. | | Deposit date: | 1998-08-03 | | Release date: | 1999-07-22 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MAD structure of Pseudomonas nautica dimeric cytochrome c552 mimicks the c4 Dihemic cytochrome domain association.
J.Mol.Biol., 289, 1999
|
|
1FWY
 
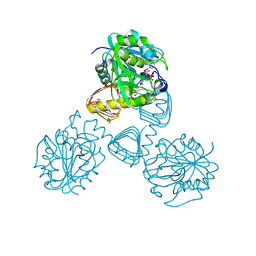 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE BOUND TO UDP-GLCNAC | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE, ... | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-25 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
2KH5
 
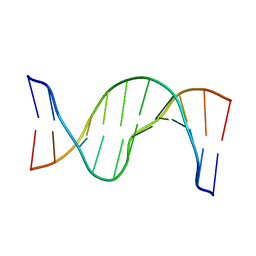 | | Solution Structure of cis-5R,6S-thymine glycol opposite complementary adenine in duplex DNA | | Descriptor: | 5'-D(*AP*CP*AP*AP*AP*CP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*GP*(CTG)P*GP*TP*TP*TP*GP*T)-3' | | Authors: | Brown, K.L. | | Deposit date: | 2009-03-24 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R,6S) thymine glycol epimer in the 5'-GTgG-3' sequence: destabilization of two base pairs at the lesion site
Nucleic Acids Res., 38, 2010
|
|
2KH6
 
 | | Solution Structure of cis-5R,6S-thymine glycol opposite complementary adenine in duplex DNA | | Descriptor: | 5'-D(*AP*CP*AP*AP*AP*CP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*GP*(CTG)P*GP*TP*TP*TP*GP*T)-3' | | Authors: | Brown, K.L. | | Deposit date: | 2009-03-24 | | Release date: | 2010-03-02 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R,6S) thymine glycol epimer in the 5'-GTgG-3' sequence: destabilization of two base pairs at the lesion site
Nucleic Acids Res., 38, 2010
|
|
1HZ0
 
 | | NMR STRUCTURE OF THE 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE (PHIP) C8-DEOXYGUANOSINE ADDUCT IN DUPLEX DNA | | Descriptor: | 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Brown, K, Cosman, M. | | Deposit date: | 2001-01-23 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1DHJ
 
 | |
1SM2
 
 | | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C.M, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-08 | | Release date: | 2004-07-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
1SNX
 
 | | CRYSTAL STRUCTURE OF APO INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
1FXJ
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-26 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
5CCP
 
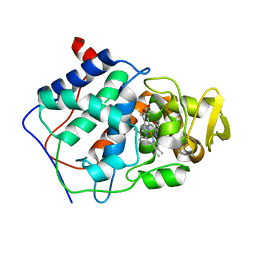 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
1QNI
 
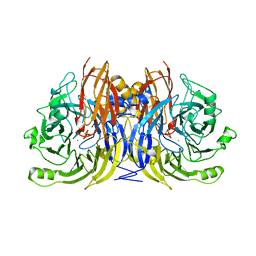 | | Crystal Structure of Nitrous Oxide Reductase from Pseudomonas nautica, at 2.4A Resolution | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 1999-10-15 | | Release date: | 2000-10-13 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel type of catalytic copper cluster in nitrous oxide reductase.
Nat.Struct.Biol., 7, 2000
|
|
2EVA
 
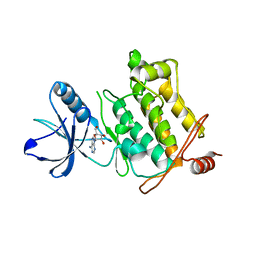 | | Structural Basis for the Interaction of TAK1 Kinase with its Activating Protein TAB1 | | Descriptor: | ADENOSINE, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Brown, K, Vial, S.C, Dedi, N, Long, J.M, Dunster, N.J, Cheetham, G.M. | | Deposit date: | 2005-10-31 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the interaction of TAK1 kinase with its activating protein TAB1
J.Mol.Biol., 354, 2005
|
|
2KH3
 
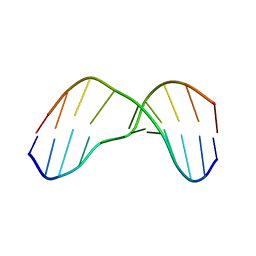 | | NMR Structure of Aflatoxin Formamidopyrimidine alpha-anomer in duplex DNA | | Descriptor: | 5'-D(*CP*TP*AP*TP*(FAG)P*AP*TP*TP*CP*A)-3', 5'-D(*TP*GP*AP*AP*TP*CP*AP*TP*AP*G)-3' | | Authors: | Brown, K.L. | | Deposit date: | 2009-03-24 | | Release date: | 2009-12-15 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Structural perturbations induced by the alpha-anomer of the aflatoxin B(1) formamidopyrimidine adduct in duplex and single-strand DNA
J.Am.Chem.Soc., 131, 2009
|
|
2KH4
 
 | | Aflatoxin Formamidopyrimidine alpha anomer in single strand DNA | | Descriptor: | 5'-D(*CP*TP*(FAG)P*A)-3' | | Authors: | Brown, K.L. | | Deposit date: | 2009-03-24 | | Release date: | 2009-12-15 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Structural perturbations induced by the alpha-anomer of the aflatoxin B(1) formamidopyrimidine adduct in duplex and single-strand DNA
J.Am.Chem.Soc., 131, 2009
|
|
2DRC
 
 | |
