7ALZ
 
 | | GqqA- a novel type of quorum quenching acylases | | 分子名称: | PHENYLALANINE, Prephenate dehydratase | | 著者 | Werner, N, Betzel, C. | | 登録日 | 2020-10-07 | | 公開日 | 2021-08-04 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | The Komagataeibacter europaeus GqqA is the prototype of a novel bifunctional N-Acyl-homoserine lactone acylase with prephenate dehydratase activity.
Sci Rep, 11, 2021
|
|
7AM0
 
 | | GqqA- a novel type of quorum quenching acylases | | 分子名称: | PHENYLALANINE, Prephenate dehydratase | | 著者 | Werner, N, Betzel, C. | | 登録日 | 2020-10-07 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Komagataeibacter europaeus GqqA is the prototype of a novel bifunctional N-Acyl-homoserine lactone acylase with prephenate dehydratase activity.
Sci Rep, 11, 2021
|
|
2QB5
 
 | | Crystal Structure of Human Inositol 1,3,4-Trisphosphate 5/6-Kinase (ITPK1) in Complex with ADP and Mn2+ | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MANGANESE (II) ION, ... | | 著者 | Chamberlain, P.P, Lesley, S.A, Spraggon, G. | | 登録日 | 2007-06-15 | | 公開日 | 2007-07-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Integration of inositol phosphate signaling pathways via human ITPK1.
J.Biol.Chem., 282, 2007
|
|
1QUZ
 
 | | Solution structure of the potassium channel scorpion toxin HSTX1 | | 分子名称: | HSTX1 TOXIN | | 著者 | Savarin, P, Romi-Lebrun, R, Zinn-Justin, S, Lebrun, B, Nakajima, T, Gilquin, B, Menez, A. | | 登録日 | 1999-07-05 | | 公開日 | 2000-07-07 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and functional consequences of the presence of a fourth disulfide bridge in the scorpion short toxins: solution structure of the potassium channel inhibitor HsTX1.
Protein Sci., 8, 1999
|
|
8OYS
 
 | |
8OYV
 
 | |
8OYX
 
 | |
8OYW
 
 | |
8OYY
 
 | |
8D8R
 
 | | SARS-CoV-2 Spike RBD in complex with DMAb 2196 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2196 heavy chain, 2196 light chain, ... | | 著者 | Du, J, Cui, J, Pallesen, J. | | 登録日 | 2022-06-08 | | 公開日 | 2022-10-19 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
8D8Q
 
 | | SARS-CoV-2 Spike RBD in complex with DMAbs 2130 and 2196 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2130 heavy chain, 2130 light chain, ... | | 著者 | Du, J, Cui, J, Pallesen, J. | | 登録日 | 2022-06-08 | | 公開日 | 2022-10-19 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
6WXJ
 
 | | CSF1R signaling is a regulator of pathogenesis in progressive MS | | 分子名称: | 4-(3-{[(2S)-2-(6-methoxypyridin-3-yl)-2,3-dihydro-1,4-benzodioxin-6-yl]methyl}-3H-imidazo[4,5-b]pyridin-6-yl)-2-methylbut-3-yn-2-amine, Macrophage colony-stimulating factor 1 receptor | | 著者 | Liu, J. | | 登録日 | 2020-05-10 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | CSF1R signaling is a regulator of pathogenesis in progressive MS.
Cell Death Dis, 11, 2020
|
|
8G0M
 
 | | Structure of complex between TV6.6 and CD98hc ECD | | 分子名称: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | 著者 | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | 登録日 | 2023-01-31 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
6W54
 
 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | 分子名称: | 4-NITROCATECHOL, COBALT (II) ION, Gallate decarboxylase, ... | | 著者 | Zeug, M, Marckovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | 登録日 | 2020-03-12 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
6WZT
 
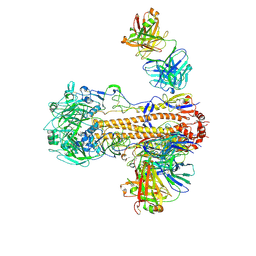 | | CryoEM structure of influenza hemagglutinin A/Victoria/361/2011 in complex with cyno antibody 3B10 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cyno antibody heavy chain, ... | | 著者 | Qiu, Y, Zhou, Y, Darricarrere, N. | | 登録日 | 2020-05-14 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
6XGC
 
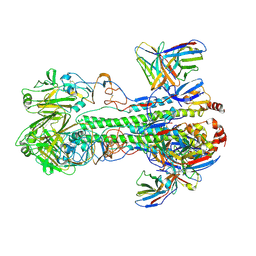 | |
6NFJ
 
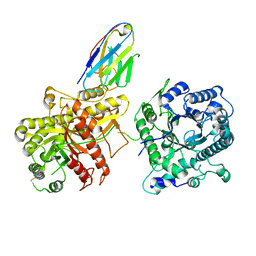 | |
6XKR
 
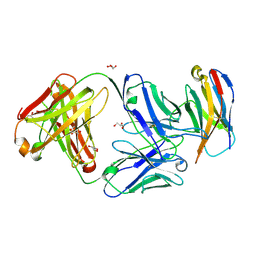 | | Structure of Sasanlimab Fab in complex with PD-1 | | 分子名称: | GLYCEROL, Programmed cell death protein 1, Sasanlimab Fab Heavy chain, ... | | 著者 | Kimberlin, C.R, Chin, S.M. | | 登録日 | 2020-06-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Pharmacologic Properties and Preclinical Activity of Sasanlimab, A High-affinity Engineered Anti-Human PD-1 Antibody.
Mol.Cancer Ther., 19, 2020
|
|
3C1C
 
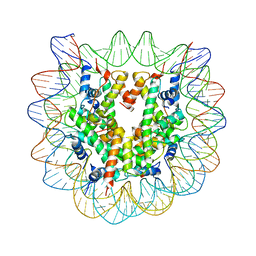 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | 分子名称: | Histone 2, H2bf, Histone H2A type 1, ... | | 著者 | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | 登録日 | 2008-01-22 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
