6N3H
 
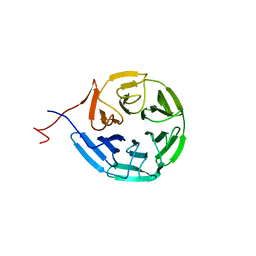 | | Crystal structure of Kelch domain of the human NS1 binding protein | | 分子名称: | Influenza virus NS1A-binding protein | | 著者 | Zhang, K, Shang, G, Padavannil, A, Fontoura, B.M.A, Chook, Y.M. | | 登録日 | 2018-11-15 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3WMC
 
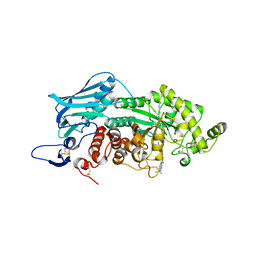 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(dimethylamino)-2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Beta-hexosaminidase | | 著者 | Chen, L, Zhou, Y, Chen, L, Yang, Q. | | 登録日 | 2013-11-16 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.095 Å) | | 主引用文献 | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
3UBX
 
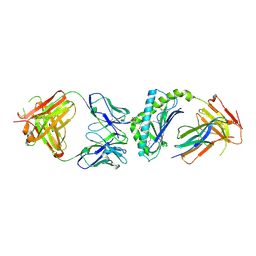 | | Crystal structure of the mouse CD1d-C20:2-aGalCer-L363 mAb Fab complex | | 分子名称: | (11Z,14Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yu, E.D, Zajonc, D.M. | | 登録日 | 2011-10-25 | | 公開日 | 2011-11-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for the recognition of C20:2-alpha GalCer by the invariant natural killer T cell receptor-like antibody L363.
J.Biol.Chem., 287, 2012
|
|
3WMB
 
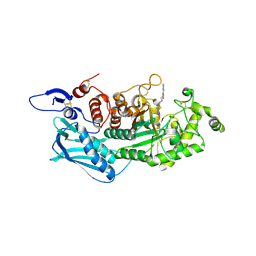 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 | | 分子名称: | 2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase | | 著者 | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | 登録日 | 2013-11-16 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
8ISO
 
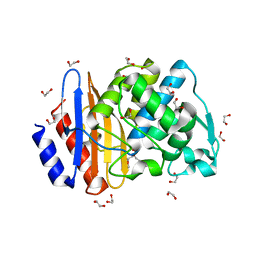 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | 分子名称: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISP
 
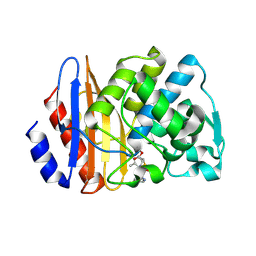 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | 分子名称: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
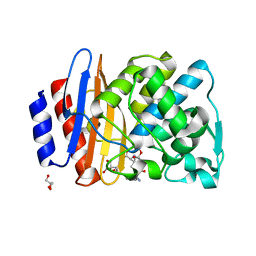 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
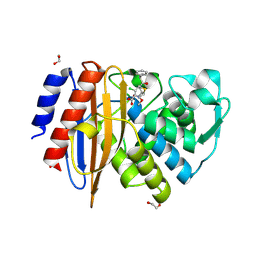 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | 分子名称: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | 登録日 | 2023-03-21 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
5YR2
 
 | |
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2023-04-11 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J1E
 
 | | AtSLAC1 in open state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2023-04-12 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8K55
 
 | |
8K57
 
 | |
5T0K
 
 | |
5T0M
 
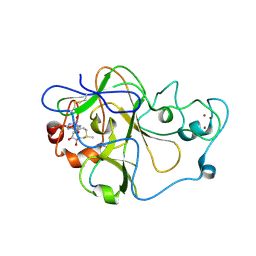 | |
5TOO
 
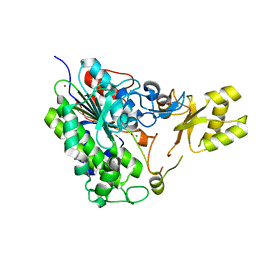 | | Crystal structure of alkaline phosphatase PafA T79S, N100A, K162A, R164A mutant | | 分子名称: | Alkaline phosphatase PafA, CHLORIDE ION, ZINC ION | | 著者 | Lyubimov, A.Y, Sunden, F, AlSadhan, I, Herschlag, D. | | 登録日 | 2016-10-18 | | 公開日 | 2017-11-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.031 Å) | | 主引用文献 | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
5O77
 
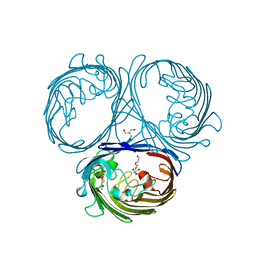 | | Klebsiella pneumoniae OmpK35 | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, OmpK35 | | 著者 | van den berg, B, Pathania, M, Zahn, M. | | 登録日 | 2017-06-08 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5O79
 
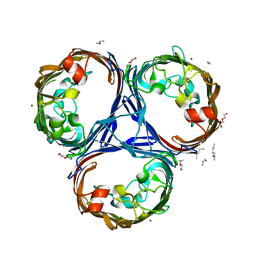 | | Klebsiella pneumoniae OmpK36 | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, OmpK36 | | 著者 | van den berg, B, Pathania, M. | | 登録日 | 2017-06-08 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5V21
 
 | |
7CHU
 
 | | Geobacillus virus E2 - ORF18 | | 分子名称: | Putative pectin lyase | | 著者 | Gong, Y. | | 登録日 | 2020-07-06 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | Structural and functional characterization of the deep-sea thermophilic bacteriophage GVE2 tailspike protein.
Int.J.Biol.Macromol., 164, 2020
|
|
2LE8
 
 | | The protein complex for DNA replication | | 分子名称: | DNA replication factor Cdt1, DNA replication licensing factor MCM6 | | 著者 | Liu, C, Wei, Z, Zhu, G. | | 登録日 | 2011-06-14 | | 公開日 | 2012-12-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into the Cdt1-mediated MCM2-7 chromatin loading
Nucleic Acids Res., 40, 2012
|
|
8T7I
 
 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | 著者 | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | 登録日 | 2023-06-20 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7G
 
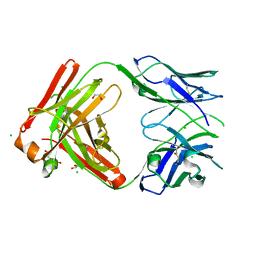 | | Structure of the CK variant of Fab F1 (FabC-F1) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | 著者 | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | 登録日 | 2023-06-20 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
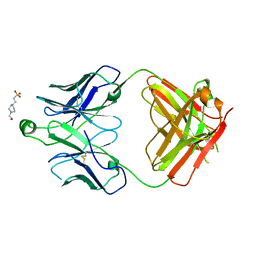 | | Structure of the S1 variant of Fab F1 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | 著者 | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | 登録日 | 2023-06-20 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
4AF3
 
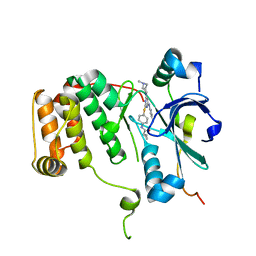 | | Human Aurora B Kinase in complex with INCENP and VX-680 | | 分子名称: | AURORA KINASE B, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, INNER CENTROMERE PROTEIN | | 著者 | Elkins, J.M, Vollmar, M, Wang, J, Picaud, S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S. | | 登録日 | 2012-01-16 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of Human Aurora B in Complex with Incenp and Vx-680.
J.Med.Chem., 55, 2012
|
|
