7VM5
 
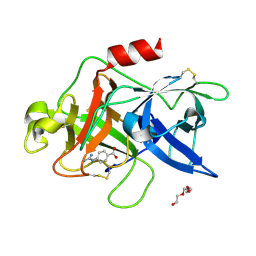 | |
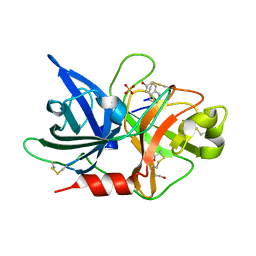
7VM6
 
 | |
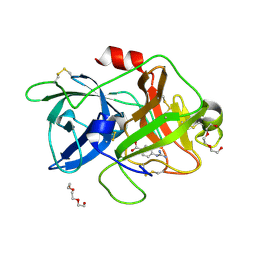
7VM4
 
 | | Crystal structure of uPA in complex with nafamostat | | 分子名称: | 4-carbamimidamidobenzoic acid, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator | | 著者 | Jiang, L.G, Huang, M.D. | | 登録日 | 2021-10-07 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
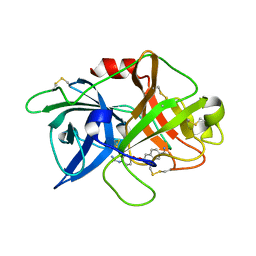
7VM7
 
 | |
5GH9
 
 | |
7WWF
 
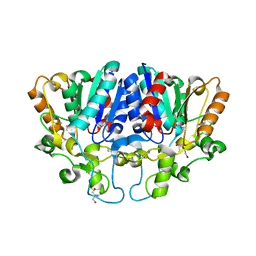 | | Crystal structure of BioH3 from Mycolicibacterium smegmatis | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Esterase | | 著者 | Yang, J, Xu, Y.C, Gan, J.H, Feng, Y.J. | | 登録日 | 2022-02-12 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-01-18 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Three enigmatic BioH isoenzymes are programmed in the early stage of mycobacterial biotin synthesis, an attractive anti-TB drug target.
Plos Pathog., 18, 2022
|
|
5GMP
 
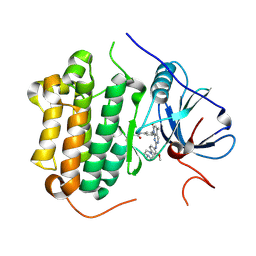 | | Crystal structure of EGFR 696-1022 T790M in complex with XTF-262 | | 分子名称: | Epidermal growth factor receptor, N-[3-[2-[[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino]-5-methyl-7-oxidanylidene-pyrido[2,3-d]pyrimidin-8-yl]phenyl]prop-2-enamide | | 著者 | Yan, X.E, Yun, C.H. | | 登録日 | 2016-07-14 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.797 Å) | | 主引用文献 | A structure-guided optimization of pyrido[2,3-d]pyrimidin-7-ones as selective inhibitors of EGFR(L858R/T790M) mutant with improved pharmacokinetic properties.
Eur J Med Chem, 126, 2017
|
|
5H7N
 
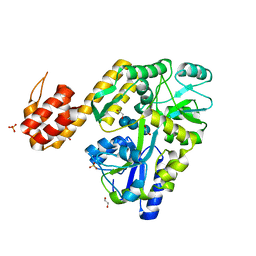 | |
5HGG
 
 | | Crystal structure of uPA in complex with a camelid-derived antibody fragment | | 分子名称: | (3S)-3-[(2S,3S,4R)-3,4-DIMETHYLTETRAHYDROFURAN-2-YL]BUTYL LAURATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Camelid Derived Antibody Fragment, ... | | 著者 | Yung, K.W.Y, Kromann-Hansen, T, Andreasen, P.A, Ngo, J.C.K. | | 登録日 | 2016-01-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2016-08-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | A Camelid-derived Antibody Fragment Targeting the Active Site of a Serine Protease Balances between Inhibitor and Substrate Behavior
J.Biol.Chem., 291, 2016
|
|
5H7Q
 
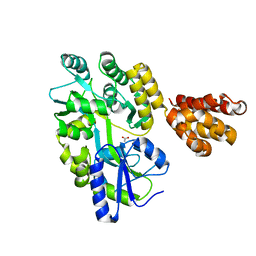 | |
5HDO
 
 | |
7U6R
 
 | |
7V0E
 
 | |
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | 著者 | Zhao, Z, Qi, J, Gao, F.G. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (two-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
