4NZW
 
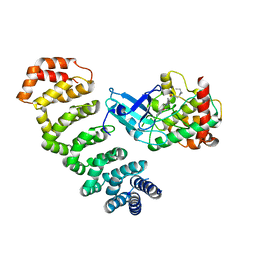 | | Crystal Structure of STK25-MO25 Complex | | 分子名称: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | 著者 | Feng, M, Hao, Q, Zhou, Z.C. | | 登録日 | 2013-12-13 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.583 Å) | | 主引用文献 | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
8JPX
 
 | |
5ZVL
 
 | |
8HNT
 
 | |
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | 分子名称: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | 著者 | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | 登録日 | 2022-12-08 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNW
 
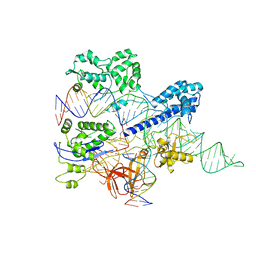 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | 著者 | Sun, W, Cheng, Z, Wang, Y. | | 登録日 | 2022-12-08 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNV
 
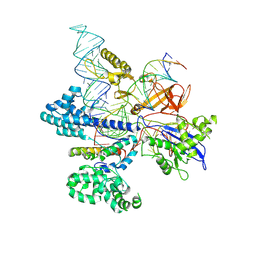 | | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | | 分子名称: | CRISPR-associated endonuclease Cas9, anti-CRISPR protein AcrIIC4, non-target strand, ... | | 著者 | Sun, W, Cheng, Z, Wang, J, Yang, X, Wang, Y. | | 登録日 | 2022-12-08 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EPX
 
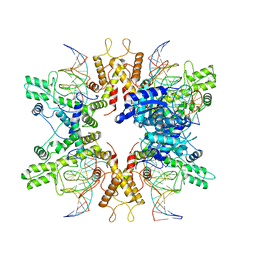 | | Type IIS Restriction Endonuclease PaqCI, DNA bound | | 分子名称: | CALCIUM ION, DNA 1a, DNA 1b, ... | | 著者 | Kennedy, M.A, Stoddard, B.L. | | 登録日 | 2022-10-06 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-06-07 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
8EM1
 
 | | Type IIS Restriction Endonuclease PaqCI, DNA Unbound | | 分子名称: | 1,2-ETHANEDIOL, PaqCI, DNA Unbound | | 著者 | Kennedy, M.A, Stoddard, B.L. | | 登録日 | 2022-09-26 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
4RAX
 
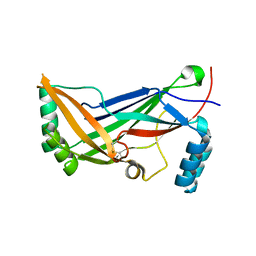 | | A regulatory domain of an ion channel | | 分子名称: | Piezo-type mechanosensitive ion channel component 1 | | 著者 | Ge, J, Yang, M. | | 登録日 | 2014-09-11 | | 公開日 | 2015-09-23 | | 最終更新日 | 2018-04-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Architecture of the mammalian mechanosensitive Piezo1 channel.
Nature, 527, 2015
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | 著者 | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | 登録日 | 2013-07-29 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.556 Å) | | 主引用文献 | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6APC
 
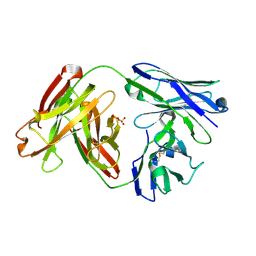 | |
6APD
 
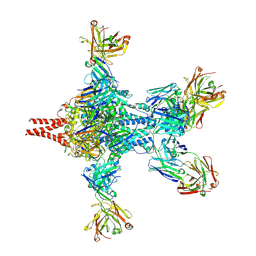 | | Crystal structure of RSV F bound by AM22 and the infant antibody ADI-19425 | | 分子名称: | AM22 Fab Heavy Chain,IGH@ protein, AM22 Fab Light Chain,Uncharacterized protein, Fusion glycoprotein F0,Envelope glycoprotein, ... | | 著者 | Wrapp, D, McLellan, J.S. | | 登録日 | 2017-08-17 | | 公開日 | 2018-03-21 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation.
Immunity, 48, 2018
|
|
6BG9
 
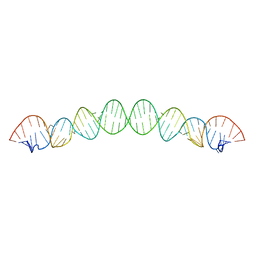 | |
8K3S
 
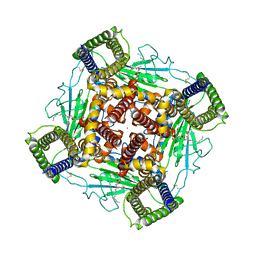 | | Structure of PKD2-F604P complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | 著者 | Chen, M.Y, Su, Q, Wang, Z.F, Yu, Y. | | 登録日 | 2023-07-16 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular and structural basis of the dual regulation of the polycystin-2 ion channel by small-molecule ligands.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6APB
 
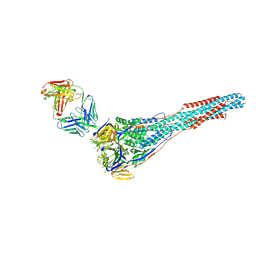 | |
4ZYK
 
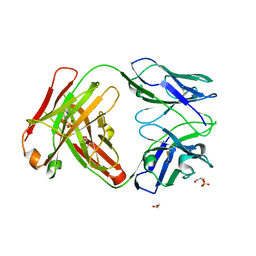 | | Crystal Structure of Quaternary-Specific RSV-Neutralizing Human Antibody AM14 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, AM14 Fab heavy chain, ... | | 著者 | Gilman, M.S.A, McLellan, J.S. | | 登録日 | 2015-05-21 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein.
Plos Pathog., 11, 2015
|
|
6NTE
 
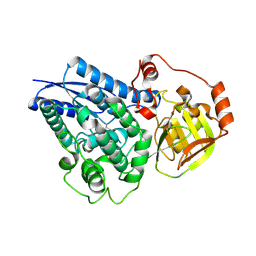 | |
4ZYP
 
 | | Crystal Structure of Motavizumab and Quaternary-Specific RSV-Neutralizing Human Antibody AM14 in Complex with Prefusion RSV F Glycoprotein | | 分子名称: | AM14 antibody Fab heavy chain, AM14 antibody light chain, Fusion glycoprotein F0,Fibritin, ... | | 著者 | Gilman, M.S.A, McLellan, J.S. | | 登録日 | 2015-05-21 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein.
Plos Pathog., 11, 2015
|
|
5H6V
 
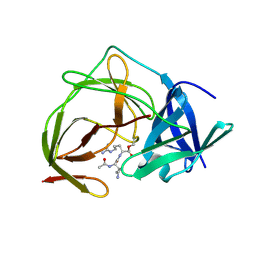 | |
6KNE
 
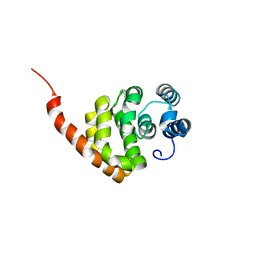 | |
6KND
 
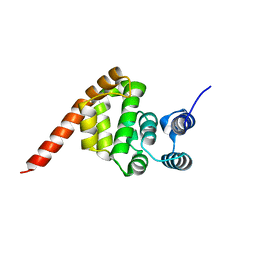 | |
4ESR
 
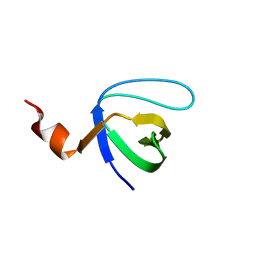 | | Molecular and Structural Characterization of the SH3 Domain of AHI-1 in Regulation of Cellular Resistance of BCR-ABL+ Chronic Myeloid Leukemia Cells to Tyrosine Kinase Inhibitors | | 分子名称: | DI(HYDROXYETHYL)ETHER, Jouberin | | 著者 | Van Petegem, X.F, Liu, P.X, Lobo, P, Jiang, X. | | 登録日 | 2012-04-23 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Molecular and structural characterization of the SH3 domain of AHI-1 in regulation of cellular resistance of BCR-ABL(+) chronic myeloid leukemia cells to tyrosine kinase inhibitors.
Proteomics, 12, 2012
|
|
5GZB
 
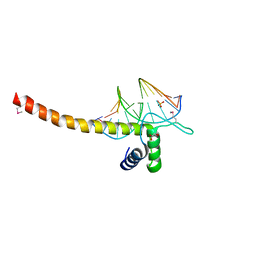 | | Crystal Structure of Transcription Factor TEAD4 in Complex with M-CAT DNA | | 分子名称: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*TP*TP*CP*CP*TP*CP*TP*C)-3'), GLYCEROL, ... | | 著者 | He, F, Shi, Z.B, Zhou, Z.C. | | 登録日 | 2016-09-28 | | 公開日 | 2017-04-19 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | DNA-binding mechanism of the Hippo pathway transcription factor TEAD4
Oncogene, 36, 2017
|
|
7C24
 
 | | Glycosidase F290Y at pH8.0 | | 分子名称: | AMMONIUM ION, GLYCEROL, Isomaltose glucohydrolase | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
