6RAA
 
 | | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003 | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Arrowsmith, C, Knapp, S, Bountra, C, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003
To Be Published
|
|
7PQS
 
 | | SRPK1 in complex with MSC2711186 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Schroeder, M, Leiendecker, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-09-20 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SRPK1 in complex with MSC2711186
To Be Published
|
|
1OJR
 
 | | L-rhamnulose-1-phosphate aldolase from Escherichia coli (mutant E192A) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Dihydroxyacetone, GLYCEROL, ... | | Authors: | Kroemer, M, Merkel, I, Schulz, G.E. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Catalytic Mechanism of L-Rhamnulose-1-Phosphate Aldolase.
Biochemistry, 42, 2003
|
|
6SFO
 
 | | MAPK14 with bound inhibitor SR-318 | | Descriptor: | 5-azanyl-~{N}-[[4-(3-cyclohexylpropylcarbamoyl)phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MAPK14 with bound inhibitor SR-318
To Be Published
|
|
1GT7
 
 | | L-rhamnulose-1-phosphate aldolase from Escherichia coli | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, RHAMNULOSE-1-PHOSPHATE ALDOLASE, ZINC ION | | Authors: | Kroemer, M, Schulz, G.E. | | Deposit date: | 2002-01-14 | | Release date: | 2002-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of L-Rhamnulose-1-Phosphate Aldolase (Class II) Solved by Low-Resolution Sir Phasing and 20-Fold Ncs Averaging
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2YGE
 
 | | E88G-N92L Mutant of N-Term HSP90 complexed with Geldanamycin | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GELDANAMYCIN, GLYCEROL | | Authors: | Roe, M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Features of the Streptomyces Hygroscopicus Htpg Reveal How Partial Geldanamycin Resistance Can Arise by Mutation to the ATP Binding Pocket of a Eukaryotic Hsp90.
Faseb J., 25, 2011
|
|
2YGF
 
 | | L89V, L93I and V136M Mutant of N-Term HSP90 complexed with Geldanamycin | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GELDANAMYCIN, GLYCEROL | | Authors: | Roe, M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Features of the Streptomyces Hygroscopicus Htpg Reveal How Partial Geldanamycin Resistance Can Arise by Mutation to the ATP Binding Pocket of a Eukaryotic Hsp90.
Faseb J., 25, 2011
|
|
3KGA
 
 | | Crystal structure of MAPKAP kinase 2 (MK2) complexed with a potent 3-aminopyrazole ATP site inhibitor | | Descriptor: | 6-{3-amino-1-[3-(1H-indol-6-yl)phenyl]-1H-pyrazol-4-yl}-3,4-dihydroisoquinolin-1(2H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Velcicky, J, Izaac, A, Be, C, Huppertz, C, Pflieger, D, Schlapbach, A, Scheufler, C. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel 3-aminopyrazole inhibitors of MK-2 discovered by scaffold hopping strategy.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2PTL
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN-BINDING DOMAIN OF PROTEIN L. COMPARISON WITH THE IGG-BINDING DOMAINS OF PROTEIN G | | Descriptor: | PROTEIN L | | Authors: | Wikstroem, M, Drakenberg, T, Forsen, S, Sjoebring, U, Bjoerck, L. | | Deposit date: | 1994-08-12 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G.
Biochemistry, 33, 1994
|
|
3M2W
 
 | | Crystal structure of MAPKAK kinase 2 (MK2) complexed with a spiroazetidine-tetracyclic ATP site inhibitor | | Descriptor: | 2'-(2-fluorophenyl)-1-methyl-6',8',9',11'-tetrahydrospiro[azetidine-3,10'-pyrido[3',4':4,5]pyrrolo[2,3-f]isoquinolin]-7'(5'H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Revesz, L, Be, C, Izaac, A, Huppertz, C, Schlapbach, A, Scheufler, C. | | Deposit date: | 2010-03-08 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | In vivo and in vitro SAR of tetracyclic MAPKAP-K2 (MK2) inhibitors. Part II.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7QGP
 
 | | STK10 (LOK) bound to Macrocycle CKJB51 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-chloranyl-2-methyl-phenyl)methyl]-3-(7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaen-5-yl)urea, CHLORIDE ION, ... | | Authors: | Berger, B.-T, Schroeder, M, Kraemer, A, Schwalm, M.P, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STK10 bound to CKJB51
To Be Published
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
3UNE
 
 | | Mouse constitutive 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UNH
 
 | | Mouse 20S immunoproteasome | | Descriptor: | CHLORIDE ION, IODIDE ION, POTASSIUM ION, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UN4
 
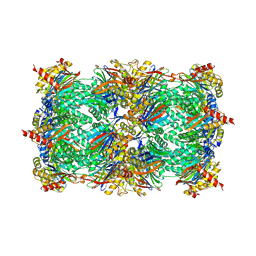 | | Yeast 20S proteasome in complex with PR-957 (morpholine) | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, Proteasome component C1, Proteasome component C11, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UNF
 
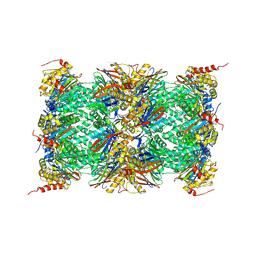 | | Mouse 20S immunoproteasome in complex with PR-957 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, CHLORIDE ION, IODIDE ION, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3UNB
 
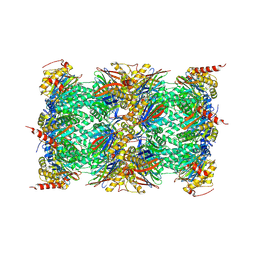 | | Mouse constitutive 20S proteasome in complex with PR-957 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
2KM6
 
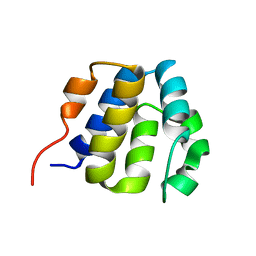 | | NMR structure of the NLRP7 Pyrin domain | | Descriptor: | NACHT, LRR and PYD domains-containing protein 7 | | Authors: | Pinheiro, A, Proell, M, Schwarzenbacher, R, Peti, W. | | Deposit date: | 2009-07-21 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity.
J.Biol.Chem., 285, 2010
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7AGA
 
 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
