3LD8
 
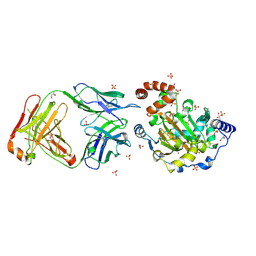 | | Structure of JMJD6 and Fab Fragments | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, GLYCEROL, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LDB
 
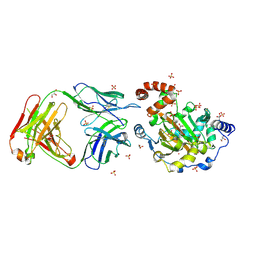 | | Structure of JMJD6 complexd with ALPHA-KETOGLUTARATE and Fab Fragment. | | Descriptor: | 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6HNF
 
 | | Structure in solution of human fibronectin type III-domain 14 | | Descriptor: | Fibronectin | | Authors: | Zhong, X, Arnolds, O, Krenczyk, O, Gajewski, J, Puetz, S, Herrmann, C, Stoll, R. | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure in Solution of Fibronectin Type III Domain 14 Reveals Its Synergistic Heparin Binding Site.
Biochemistry, 57, 2018
|
|
3CQ8
 
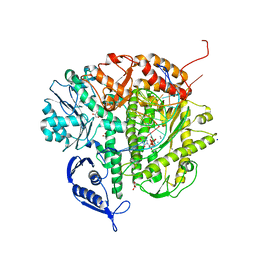 | | Ternary complex of the L415F mutant RB69 exo(-)polymerase | | Descriptor: | CALCIUM ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*DCP*DC)-3'), ... | | Authors: | Zhong, X, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2008-04-02 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a replicative DNA polymerase mutant with reduced fidelity and increased translesion synthesis capacity.
Nucleic Acids Res., 36, 2008
|
|
8GTJ
 
 | |
8GTK
 
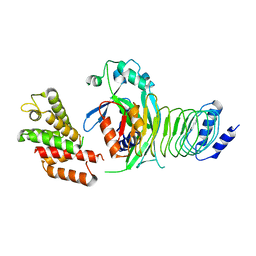 | |
3WYD
 
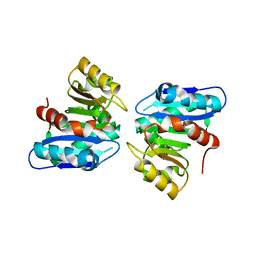 | |
7T0R
 
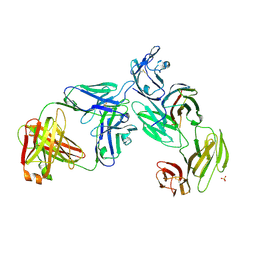 | | Crystal structure of the anti-CD4 adnectin 6940_B01 as a complex with the extracellular domains of CD4 and ibalizumab fAb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adnectin 6940_B01, Ibalizumab Heavy Chain, ... | | Authors: | Williams, S.P, Concha, N.O, Wensel, D.L, Hong, X. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Novel Bent Conformation of CD4 Induced by HIV-1 Inhibitor Indirectly Prevents Productive Viral Attachment.
J.Mol.Biol., 434, 2021
|
|
1NM7
 
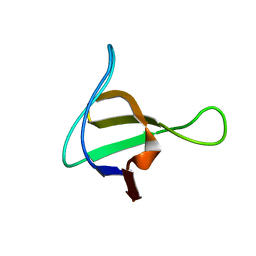 | | Solution structure of the ScPex13p SH3 domain | | Descriptor: | Peroxisomal Membrane Protein PAS20 | | Authors: | Pires, J.R, Hong, X, Brockmann, C, Volkmer-Engert, R, Schneider-Mergener, J, Oschkinat, H, Erdmann, R. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The ScPex13p SH3 Domain Exposes Two Distinct Binding Sites for Pex5p and Pex14p
J.Mol.Biol., 326, 2003
|
|
2PXJ
 
 | | The complex structure of JMJD2A and monomethylated H3K36 peptide | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, ... | | Authors: | Chen, Z, Zang, J, Kappler, J, Hong, X, Crawford, F, Zhang, G. | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2P5B
 
 | | The complex structure of JMJD2A and trimethylated H3K36 peptide | | Descriptor: | FE (II) ION, Histone H3, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Zhang, G, Chen, Z, Zang, J, Hong, X, Shi, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2007-06-12 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2GP5
 
 | | Crystal structure of catalytic core domain of jmjd2A complexed with alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Jumonji domain-containing protein 2A, ... | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
2GP3
 
 | | Crystal structure of the catalytic core domain of jmjd2a | | Descriptor: | FE (II) ION, Jumonji domain-containing protein 2A, ZINC ION | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
3O2D
 
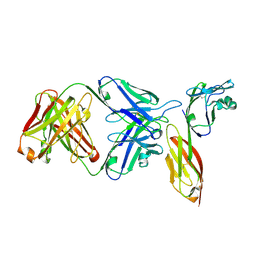 | | Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody | | Descriptor: | T-cell surface glycoprotein CD4, ibalizumab heavy chain, ibalizumab light chain | | Authors: | Freeman, M.M, Seaman, M.S, Rits-Volloch, S, Hong, X, Ho, D.D, Chen, B. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of HIV-1 Primary Receptor CD4 in Complex with a Potent Antiviral Antibody.
Structure, 18, 2010
|
|
3PYY
 
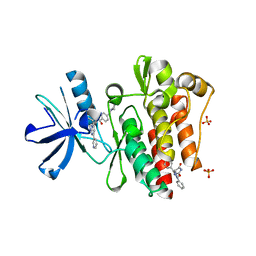 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
1HUZ
 
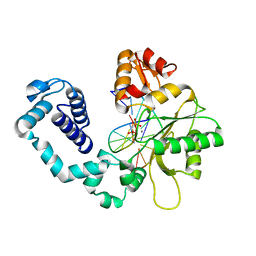 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1HUO
 
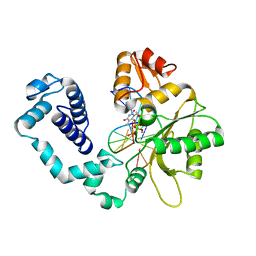 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
4QD2
 
 | | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex | | Descriptor: | CALCIUM ION, Cadherin-1, Hemagglutinin component HA17, ... | | Authors: | Lee, K, Zhong, X, Gu, S, Kruel, A, Dorner, M.B, Perry, K, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex.
Science, 344, 2014
|
|
6VIJ
 
 | | Crystal structure of mouse RABL3 in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rab-like protein 3 | | Authors: | Su, L, Tomchick, D.R, Beutler, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetic and structural studies of RABL3 reveal an essential role in lymphoid development and function.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VII
 
 | |
6VIK
 
 | |
6VIH
 
 | | The ligand-free structure of mouse RABL3 | | Descriptor: | Rab-like protein 3 | | Authors: | Su, L, Tomchick, D.R, Beutler, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Genetic and structural studies of RABL3 reveal an essential role in lymphoid development and function.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4ONJ
 
 | | Crystal structure of the catalytic domain of ntDRM | | Descriptor: | DNA methyltransferase, SINEFUNGIN | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular Mechanism of Action of Plant DRM De Novo DNA Methyltransferases.
Cell(Cambridge,Mass.), 157, 2014
|
|
4ONQ
 
 | |
8GTN
 
 | |
