3WCD
 
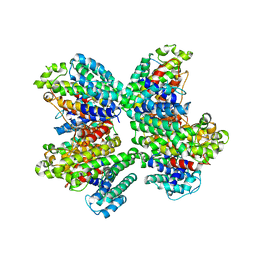 | | The complex structure of HsSQS wtih ligand, WC-9 | | Descriptor: | 2-(4-phenoxyphenoxy)ethyl thiocyanate, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCJ
 
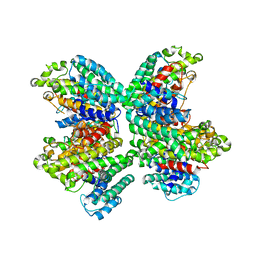 | | The complex structure of HsSQS wtih ligand,E5700 | | Descriptor: | (3R)-3-({2-benzyl-6-[(3R,4S)-3-hydroxy-4-methoxypyrrolidin-1-yl]pyridin-3-yl}ethynyl)-1-azabicyclo[2.2.2]octan-3-ol, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCF
 
 | | The complex structure of HsSQS wtih ligand,BPH1218 | | Descriptor: | Squalene synthase, hydrogen [(1S)-2-(3-decyl-1H-imidazol-3-ium-1-yl)-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
8ZQE
 
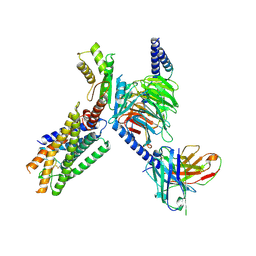 | | Cryo-EM structure of the GPR15L(C11)-bound GPR15 complex | | Descriptor: | GPR15, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Z.Y, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-02 | | Release date: | 2024-07-10 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular recognition of the atypical chemokine-like peptide GPR15L by its cognate receptor GPR15.
Cell Discov, 10, 2024
|
|
8YUV
 
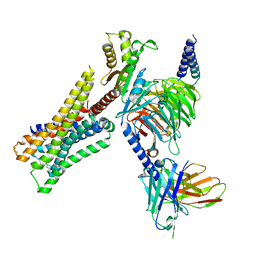 | | Cryo-EM structure of the immepip-bound H3R-Gi complex | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUT
 
 | | Cryo-EM structure of the amthamine-bound H2R-Gs complex | | Descriptor: | 5-(2-azanylethyl)-4-methyl-1,3-thiazol-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUU
 
 | | Cryo-EM structure of the histamine-bound H3R-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
7ULU
 
 | | Human DDAH1 soaked with its inhibitor ClPyrAA | | Descriptor: | (2S)-2-amino-4-[(pyridin-2-yl)amino]butanoic acid, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Butrin, A, Zheng, Y, Tuley, A, Liu, D, Fast, W. | | Deposit date: | 2022-04-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of a switchable electrophile fragment into a potent and selective covalent inhibitor of human DDAH1
To Be Published
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
6JAT
 
 | | Crystal structure of SETD3 bound to Actin peptide and SFG | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Actin, gamma-enteric smooth muscle, ... | | Authors: | Li, H, Zheng, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Crystal structure of SETD3 bound to Actin peptide and SFG
to be published
|
|
8ZSP
 
 | | Cryo-EM structure of the LSD-bound hTAAR1-Gs complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8ZSV
 
 | | Cryo-EM structure of the RO5263397-bound mTAAR1-Gs complex | | Descriptor: | (4~{S})-4-(3-fluoranyl-2-methyl-phenyl)-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8ZSS
 
 | | Cryo-EM structure of the RO5263397-bound hTAAR1-Gs complex | | Descriptor: | (4~{S})-4-(3-fluoranyl-2-methyl-phenyl)-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8ZSJ
 
 | | Cryo-EM structure of the apo hTAAR1-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
3S29
 
 | |
1G72
 
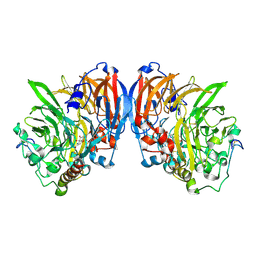 | | CATALYTIC MECHANISM OF QUINOPROTEIN METHANOL DEHYDROGENASE: A THEORETICAL AND X-RAY CRYSTALLOGRAPHIC INVESTIGATION | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE HEAVY SUBUNIT, METHANOL DEHYDROGENASE LIGHT SUBUNIT, ... | | Authors: | Zheng, Y, Xia, Z, Chen, Z, Bruice, T.C, Mathews, F.S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of quinoprotein methanol dehydrogenase: A theoretical and x-ray crystallographic investigation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6KXB
 
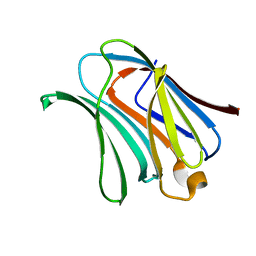 | | Galectin-3 CRD binds to GalA trimer | | Descriptor: | Galectin-3, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid | | Authors: | Su, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Topsy-turvy binding of negatively charged homogalacturonan oligosaccharides to galectin-3.
Glycobiology, 31, 2021
|
|
5I4V
 
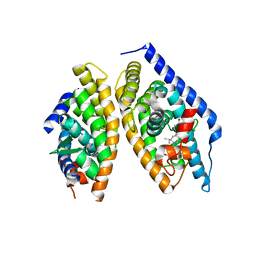 | | Discovery of novel, orally efficacious Liver X Receptor (LXR) beta agonists | | Descriptor: | Oxysterols receptor LXR-beta,Nuclear receptor coactivator 2, Retinoic acid receptor RXR-beta,Nuclear receptor coactivator 2, {2-[(2R)-4-[4-(hydroxymethyl)-3-(methylsulfonyl)phenyl]-2-(propan-2-yl)piperazin-1-yl]-4-(trifluoromethyl)pyrimidin-5-yl}methanol | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of a Novel, Orally Efficacious Liver X Receptor (LXR) beta Agonist.
J.Med.Chem., 59, 2016
|
|
8HCI
 
 | |
8HI8
 
 | |
8HI7
 
 | |
7ESC
 
 | | FmnB complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7ESA
 
 | | the complex structure of flavin transferase FmnB complexed with FAD | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
