7YPF
 
 | |
4RFR
 
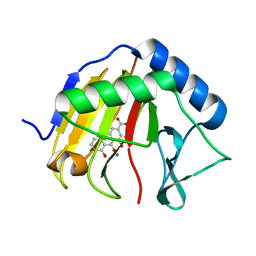 | | Complex structure of AlkB/rhein | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase AlkB, MANGANESE (II) ION | | Authors: | Li, Q, Huang, Y, Li, J.F, Yang, C.-G. | | Deposit date: | 2014-09-27 | | Release date: | 2016-04-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rhein Inhibits AlkB Repair Enzymes and Sensitizes Cells to Methylated DNA Damage.
J.Biol.Chem., 291, 2016
|
|
2DD8
 
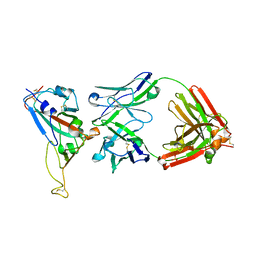 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGG Heavy Chain, IGG Light Chain, ... | | Authors: | Prabakaran, P, Gan, J.H, Feng, Y, Zhu, Z.Y, Xiao, X.D, Ji, X, Dimitrov, D.S. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Severe Acute Respiratory Syndrome Coronavirus Receptor-binding Domain Complexed with Neutralizing Antibody
J.Biol.Chem., 281, 2006
|
|
7C23
 
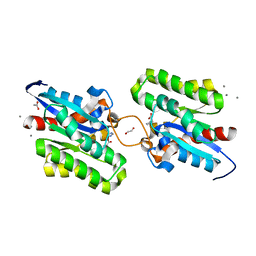 | | Crystal structure of CrmE10, a SGNH-hydrolase family esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7C29
 
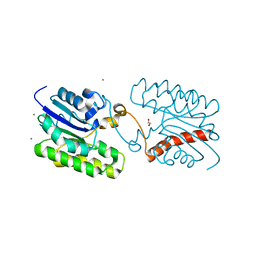 | | Esterase CrmE10 mutant-D178A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7BP6
 
 | |
7BP2
 
 | |
7BP5
 
 | |
7BP4
 
 | |
7WWF
 
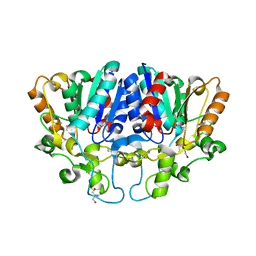 | | Crystal structure of BioH3 from Mycolicibacterium smegmatis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Esterase | | Authors: | Yang, J, Xu, Y.C, Gan, J.H, Feng, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-06 | | Last modified: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Three enigmatic BioH isoenzymes are programmed in the early stage of mycobacterial biotin synthesis, an attractive anti-TB drug target.
Plos Pathog., 18, 2022
|
|
5XM9
 
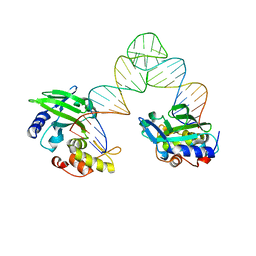 | |
5XMA
 
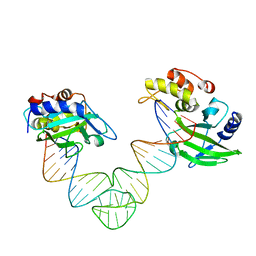 | |
7WID
 
 | | Crystal structure of Staphylococcus aureus ClpP in complex with ZG180 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2022-01-03 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7WGS
 
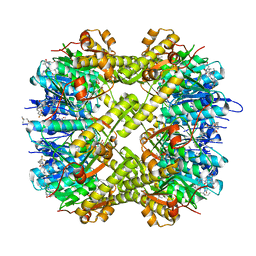 | | Structure of ClpP from Staphylococcus aureus in complex with (S)-ZG197 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2R)-butan-2-yl]-8-[(1S)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
7WH5
 
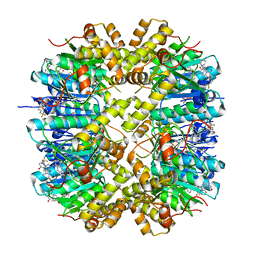 | | Crystal structure of human ClpP in complex with ZG180 | | Descriptor: | (6S,9aS)-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-12-29 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
5XM8
 
 | |
5ZFZ
 
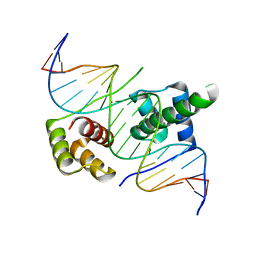 | | Crystal structure of human DUX4 homeodomains bound to A12T DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*TP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*AP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5ZFW
 
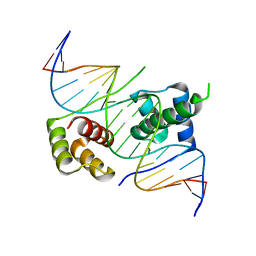 | | Crystal structure of human DUX4 homeodomains bound to A11G DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*GP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*CP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5ZFY
 
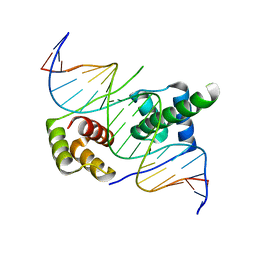 | | Crystal structure of human DUX4 homeodomains bound to A12C DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*CP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*GP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5Z6Z
 
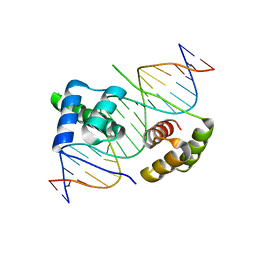 | | Crystal structure of human DUX4 homeodomains bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-01-26 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
6IMJ
 
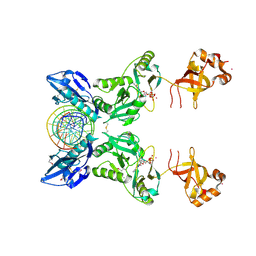 | | The crystal structure of Se-AsfvLIG:DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
6IMK
 
 | | The crystal structure of AsfvLIG:CG complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*G)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
6IML
 
 | | The crystal structure of AsfvLIG:CT1 complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
6KHY
 
 | | The crystal structure of AsfvAP:AG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
6IMN
 
 | | The crystal structure of AsfvLIG:CT2 complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*CP*CP*GP*CP*AP*TP*CP*CP*CP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*GP*TP*CP*GP*GP*AP*CP*TP*GP*G)-3'), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Nat Commun, 10, 2019
|
|
