4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
3E87
 
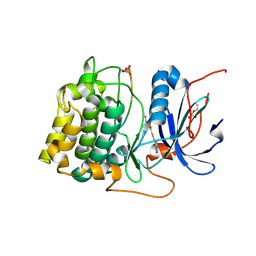 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | Glycogen synthase kinase-3 beta peptide, N-[(1S)-2-amino-1-phenylethyl]-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)thiophene-2-carboxamide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E8E
 
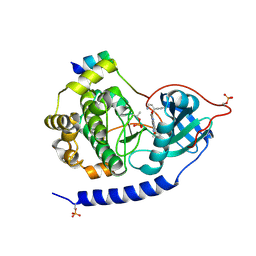 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, PKI inhibitor peptide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4I8X
 
 | | Crystal structure of rabbit LDHA in complex with AP27460 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4I9N
 
 | | Crystal structure of rabbit LDHA in complex with AP28161 and AP28122 | | Descriptor: | 6-({2-[(5-chloro-4-{[(2S)-2,3-dihydroxypropyl]oxy}-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, 6-[3-(carboxymethoxy)-5-fluorophenyl]pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
8TBF
 
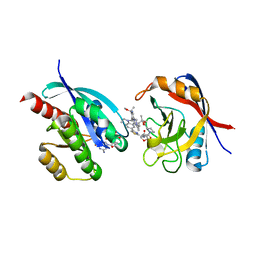 | | Tricomplex of RMC-7977, KRAS WT, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
7CXS
 
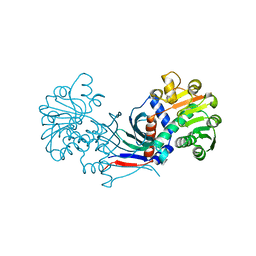 | |
7CXV
 
 | | Crystal structure of CmnK | | Descriptor: | CmnK | | Authors: | Chang, C.Y, Hsu, S.H. | | Deposit date: | 2020-09-02 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of Enzymes Catalyzing the Formation of the Nonproteinogenic Amino Acid l-Dap in Capreomycin Biosynthesis.
Biochemistry, 60, 2021
|
|
7CXU
 
 | | Crystal structure of CmnK in complex with NAD+ | | Descriptor: | CmnK, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chang, C.Y, Hsu, S.H. | | Deposit date: | 2020-09-02 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of Enzymes Catalyzing the Formation of the Nonproteinogenic Amino Acid l-Dap in Capreomycin Biosynthesis.
Biochemistry, 60, 2021
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
1F9I
 
 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT Y42F | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-10 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
1F98
 
 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT T50V | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-07 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
7SN0
 
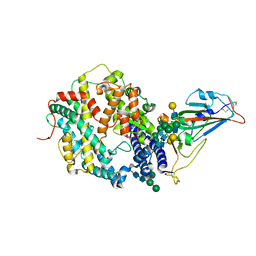 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Clark, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN1
 
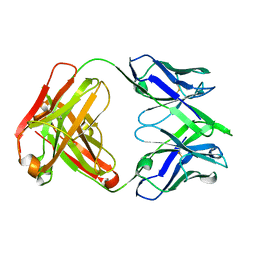 | |
7QWR
 
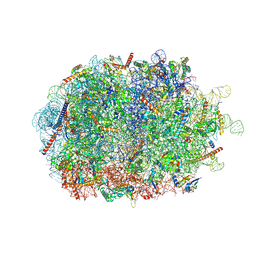 | | Structure of the ribosome-nascent chain containing an ER signal sequence in complex with NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7QWS
 
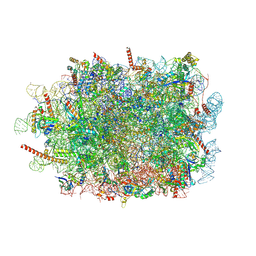 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7QWQ
 
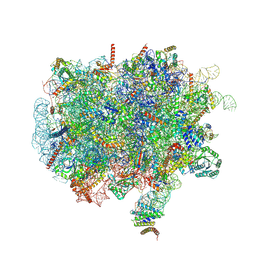 | | Ternary complex of ribosome nascent chain with SRP and NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
6EYF
 
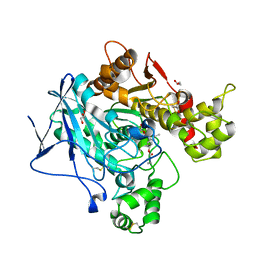 | | Butyrylcolinesterase expressed in CHO cells co-crystallised with a rivastigmine analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, GLYCEROL, ... | | Authors: | Brazzolotto, X, De la Mora, E, Dighe, S, Ross, B. | | Deposit date: | 2017-11-12 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rivastigmine and metabolite analogues with putative Alzheimer's disease-modifying properties in a Caenorhabditis elegans model
Commun Chem, 2019
|
|
7QAZ
 
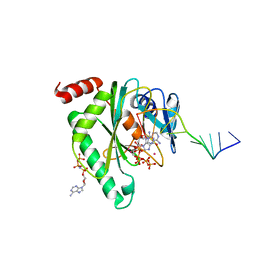 | |
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | Descriptor: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,YL-365 | | Authors: | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5DH3
 
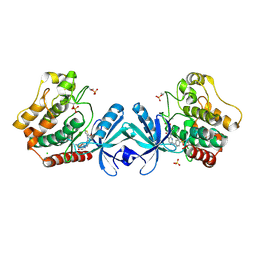 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
7QVH
 
 | |
5TDK
 
 | | RNA decamer duplex with eight 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
