9CLW
 
 | | Cryo-EM structure of Gq-coupled FFA2 in complex with TUG-1375 and 4-CMTB | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, (2~{S})-2-(4-chlorophenyl)-3-methyl-~{N}-(1,3-thiazol-2-yl)butanamide, CHOLESTEROL, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2024-07-12 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Allosteric modulation and biased signalling at free fatty acid receptor 2.
Nature, 2025
|
|
9CM7
 
 | | Cryo-EM structure of Gi-coupled FFA2 in complex with TUG-1375 and AZ-1729 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2024-07-12 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Allosteric modulation and biased signalling at free fatty acid receptor 2.
Nature, 2025
|
|
9CM3
 
 | | Cryo-EM structure of Gq-coupled FFA2 in complex with TUG-1375 and compound 187 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, 4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-7-(2-fluorobenzene-1-sulfonyl)-2-methyl-5H-pyrrolo[3,2-d]pyrimidin-6-amine, CHOLESTEROL, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2024-07-12 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Allosteric modulation and biased signalling at free fatty acid receptor 2.
Nature, 2025
|
|
8T3V
 
 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | Descriptor: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3O
 
 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3Q
 
 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3S
 
 | | Cryo-EM structure of the Butyrate bound FFA2-Gq complex | | Descriptor: | CHOLESTEROL, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8SG1
 
 | | Cryo-EM structure of CMKLR1 signaling complex | | Descriptor: | CHOLESTEROL, Chemerin 9, Chemerin-like receptor 1, ... | | Authors: | Zhang, X, Zhang, C. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of G protein-Coupled receptor CMKLR1 activation and signaling induced by a chemerin-derived agonist.
Plos Biol., 21, 2023
|
|
9JUY
 
 | | X-ray crystal structure of Y16513 in CBP | | Descriptor: | (5~{S})-1-(3-chloranyl-4-methoxy-phenyl)-5-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]pyrrolidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
9JUT
 
 | | X-ray crystal structure of Y16524 in EP300 | | Descriptor: | (6~{S})-1-(3-chloranyl-4-methoxy-phenyl)-6-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]piperidin-2-one, Histone acetyltransferase p300 | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
9JUU
 
 | | X-ray crystal structure of Y16515 in CBP | | Descriptor: | (5~{S})-1-(3-chloranyl-4-methoxy-phenyl)-5-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{R})-2-morpholin-4-ylpropyl]imidazol-2-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
9J6O
 
 | | Crystal Structure of bromodomain of human CBP in complex with the inhibitor CZL-046 | | Descriptor: | (3~{S},5~{S})-1-[3,4-bis(fluoranyl)phenyl]-5-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]-3-fluoranyl-pyrrolidin-2-one, CHLORIDE ION, CREB-binding protein | | Authors: | Zhang, Y, Zhang, C, Chen, Z, Yang, H, Huang, X, Li, Y, Xu, Y. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of CZL-046 with an ( S )-3-Fluoropyrrolidin-2-one Scaffold as a p300 Bromodomain Inhibitor for the Treatment of Multiple Myeloma.
J.Med.Chem., 67, 2024
|
|
7JVB
 
 | | Crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) with nanobody Nb20 | | Descriptor: | CACODYLATE ION, Nanobody Nb20, Spike protein S1 | | Authors: | Xiang, Y, Xiao, Z, Liu, H, Sang, Z, Schneidman-Duhovny, D, Zhang, C, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science, 370, 2020
|
|
2LSR
 
 | | Solution structure of harmonin N terminal domain in complex with a exon68 encoded peptide of cadherin23 | | Descriptor: | Harmonin, peptide from Cadherin-23 | | Authors: | Pan, L, Wu, L, Zhang, C, Zhang, M. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Large protein assemblies formed by multivalent interactions between cadherin23 and harmonin suggest a stable anchorage structure at the tip link of stereocilia.
J.Biol.Chem., 287, 2012
|
|
7Y36
 
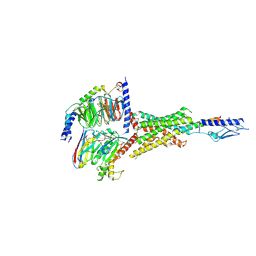 | | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-10 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex
to be published
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT7
 
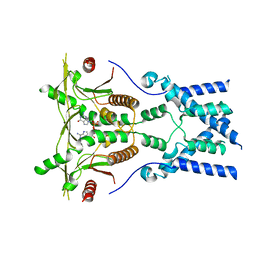 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound dimeric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT8
 
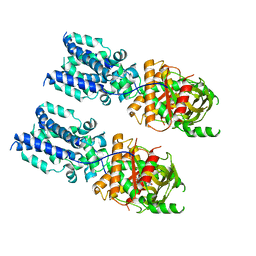 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound tetrameric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT6
 
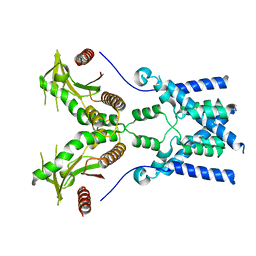 | | Cryo-EM structure of full-length chicken STING in the apo state | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
