1OHU
 
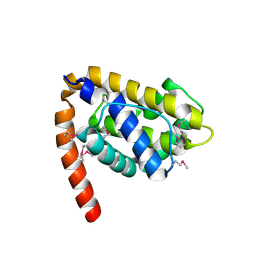 | | Structure of Caenorhabditis elegans CED-9 | | Descriptor: | APOPTOSIS REGULATOR CED-9 | | Authors: | Jeong, J.-S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Unique Structural Features of a Bcl-2 Family Protein Ced-9 and Biophysical Characterization of Ced-9/Egl-1 Interactions
Cell Death Differ., 10, 2003
|
|
7SZR
 
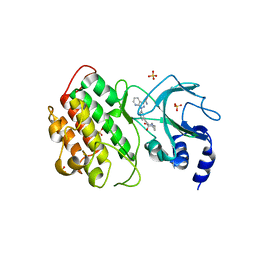 | | NIK bound to inhibitor G02792917 | | Descriptor: | 1-(3-{[(1R,4R,5S)-4-hydroxy-2-methyl-3-oxo-2-azabicyclo[3.1.0]hexan-4-yl]ethynyl}phenyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Liau, N.P.D, Hymowitz, S.G. | | Deposit date: | 2021-11-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Filling a nick in NIK: Extending the half-life of a NIK inhibitor through structure-based drug design.
Bioorg.Med.Chem.Lett., 89, 2023
|
|
1E3V
 
 | | Crystal structure of ketosteroid isomerase from Psedomonas putida complexed with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Kim, J.-S, Oh, B.-H. | | Deposit date: | 2000-06-24 | | Release date: | 2001-03-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1E3R
 
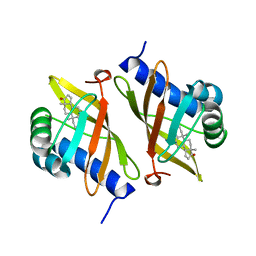 | | Crystal structure of ketosteroid isomerase mutant D40N (D38N TI numbering) from Pseudomonas putida complexed with androsten-3beta-ol-17-one | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Hyun, B.-H, Oh, B.-H. | | Deposit date: | 2000-06-22 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1OGX
 
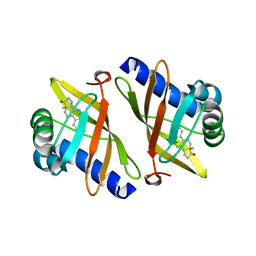 | | High Resolution Crystal Structure Of Ketosteroid Isomerase Mutant D40N(D38N, Ti Numbering) from Pseudomonas putida Complexed With Equilenin At 2.0 A Resolution. | | Descriptor: | EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-05-17 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbation of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1RCS
 
 | | NMR STUDY OF TRP REPRESSOR-OPERATOR DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*AP*CP*G)-3'), TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the trp repressor-operator DNA complex.
J.Mol.Biol., 238, 1994
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | Descriptor: | M156R | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-07-05 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
4PPZ
 
 | | Crystal structure of zinc-bound succinyl-diaminopimelate desuccinylase from Neisseria meningitidis MC58 | | Descriptor: | PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
5VFI
 
 | | Bruton's tyrosine kinase (BTK) with GDC-0853 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3'-(hydroxymethyl)-1-methyl-5-({5-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-2-yl}amino)-6-oxo[1,6-dihydro[3,4'-bipyridine]]-2'-yl]-7,7-dimethyl-3,4,7,8-tetrahydro-2H-cyclopenta[4,5]pyrrolo[1,2-a]pyrazin-1(6H)-one, SULFATE ION, ... | | Authors: | Steinbacher, S, Eigenbrot, C. | | Deposit date: | 2017-04-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of GDC-0853: A Potent, Selective, and Noncovalent Bruton's Tyrosine Kinase Inhibitor in Early Clinical Development.
J. Med. Chem., 61, 2018
|
|
5EJJ
 
 | | Crystal structure of UfSP from C.elegans | | Descriptor: | Ufm1-specific protease | | Authors: | Kim, K, Ha, B, Kim, E.E. | | Deposit date: | 2015-11-02 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
5G2C
 
 | | The crystal structure of light-driven chloride pump ClR (T102D) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
4QMG
 
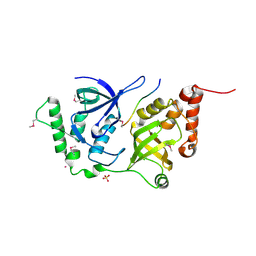 | | The Structure of MTDH-SND1 Complex Reveals Novel Cancer-Promoting Interactions | | Descriptor: | CESIUM ION, GLYCEROL, Protein LYRIC, ... | | Authors: | Guo, F, Stanevich, V, Wan, L, Satyshur, K, Kang, Y, Xing, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Insights into the Tumor-Promoting Function of the MTDH-SND1 Complex.
Cell Rep, 8, 2014
|
|
4O23
 
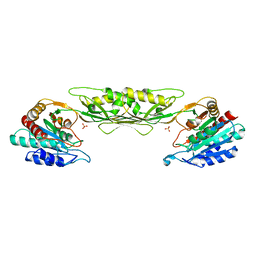 | | Crystal structure of mono-zinc form of succinyl diaminopimelate desuccinylase from Neisseria meningitidis MC58 | | Descriptor: | SULFATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
4PQA
 
 | | Crystal Structure of succinyl-diaminopimelate desuccinylase from Neisseria meningitidis MC58 in complex with the Inhibitor Captopril | | Descriptor: | L-CAPTOPRIL, SULFATE ION, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Nocek, B, Starus, A, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
5G54
 
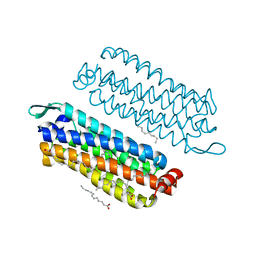 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2A
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0 with Bromide ion. | | Descriptor: | BROMIDE ION, CHLORIDE PUMPING RHODOPSIN, RETINAL | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
6DTX
 
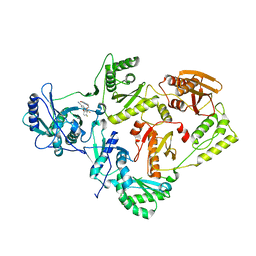 | | Wildtype HIV-1 Reverse Transcriptase in complex with JLJ 578 | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, GLYCEROL, Reverse transcriptase/ribonuclease H, ... | | Authors: | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.327 Å) | | Cite: | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6DTW
 
 | | HIV-1 Reverse Transcriptase Y181C Mutant in complex with JLJ 578 | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2PE0
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1 (PDK1) 5-Hydroxy-3-[1-(1H-pyrrol-2-yl)-eth-(Z)-ylidene]-1,3-dihydro-indol-2-one COMPLEX | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 5-HYDROXY-3-[(1R)-1-(1H-PYRROL-2-YL)ETHYL]-2H-INDOL-2-ONE, GLYCEROL, ... | | Authors: | Whitlow, M, Adler, M. | | Deposit date: | 2007-04-01 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Indolinone based phosphoinositide-dependent kinase-1 (PDK1) inhibitors. Part 1: Design, synthesis and biological activity.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3TGM
 
 | | X-Ray Crystal Structure of Human Heme Oxygenase-1 in Complex with 1-(1H-imidazol-1-yl)-4,4-diphenyl-2 butanone | | Descriptor: | 1-(1H-imidazol-1-yl)-4,4-diphenylbutan-2-one, HEXANE-1,6-DIOL, Heme oxygenase 1, ... | | Authors: | Rahman, M.N, Jia, Z. | | Deposit date: | 2011-08-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A novel, "double-clamp" binding mode for human heme oxygenase-1 inhibition.
Plos One, 7, 2012
|
|
4EHE
 
 | | B-Raf Kinase Domain in Complex with an Aminothienopyrimidine-based Inhibitor | | Descriptor: | 4-amino-N-{2,6-difluoro-3-[(propylsulfonyl)amino]phenyl}thieno[3,2-d]pyrimidine-7-carboxamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C. | | Deposit date: | 2012-04-02 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Potent and selective aminopyrimidine-based B-raf inhibitors with favorable physicochemical and pharmacokinetic properties.
J.Med.Chem., 55, 2012
|
|
4EHG
 
 | | B-Raf Kinase Domain in Complex with an Aminopyridimine-based Inhibitor | | Descriptor: | N-{2,4-difluoro-3-[({6-[(2-hydroxyethyl)amino]pyrimidin-4-yl}carbamoyl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C. | | Deposit date: | 2012-04-02 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent and selective aminopyrimidine-based B-raf inhibitors with favorable physicochemical and pharmacokinetic properties.
J.Med.Chem., 55, 2012
|
|
7WJ5
 
 | | Cryo-EM structure of human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand binding specificity | | Descriptor: | Gai1 antibody (scfv16), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Heo, Y.S, Yoon, E.J, Jeon, Y.E, Yun, J.-H, Ishimoto, N, Woo, H, Park, S.Y, Song, J, Lee, W.T. | | Deposit date: | 2022-01-05 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of the human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand-binding specificity.
Elife, 11, 2022
|
|
