1UP1
 
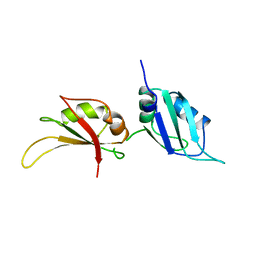 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Xu, R.-M, Jokhan, L, Cheng, X, Mayeda, A, Krainer, A.R. | | Deposit date: | 1997-03-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs.
Structure, 5, 1997
|
|
6MBP
 
 | | GLP Methyltransferase with Inhibitor EML741- P3121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
10MH
 
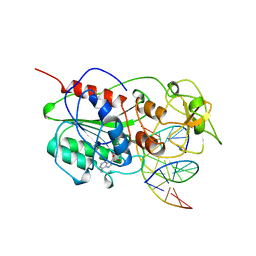 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
16VP
 
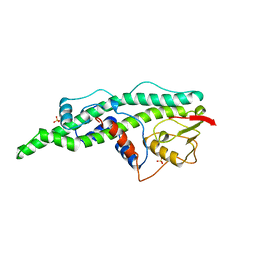 | | CONSERVED CORE OF THE HERPES SIMPLEX VIRUS TRANSCRIPTIONAL REGULATORY PROTEIN VP16 | | Descriptor: | PROTEIN (VP16, VMW65, ATIF), ... | | Authors: | Liu, Y, Gong, W, Huang, C.C, Herr, W, Cheng, X. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16.
Genes Dev., 13, 1999
|
|
1BM8
 
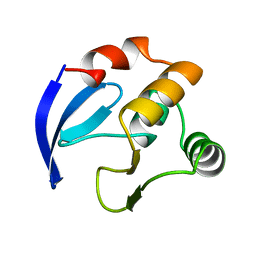 | | DNA-BINDING DOMAIN OF MBP1 | | Descriptor: | TRANSCRIPTION FACTOR MBP1 | | Authors: | Xu, R.-M, Koch, C, Liu, Y, Horton, J.R, Knapp, D, Nasmyth, K, Cheng, X. | | Deposit date: | 1998-07-29 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis.
Structure, 5, 1997
|
|
7JVQ
 
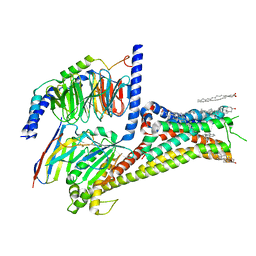 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVP
 
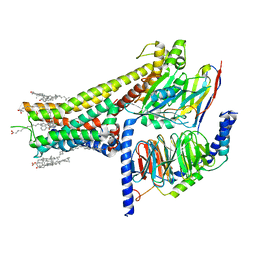 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
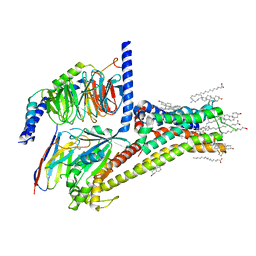 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVR
 
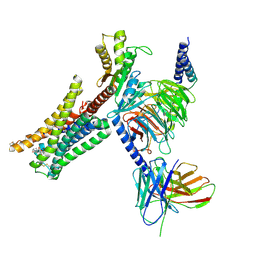 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
3SVM
 
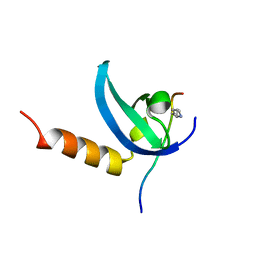 | | Human MPP8 - human DNMT3AK47me2 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2011-07-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
3SWC
 
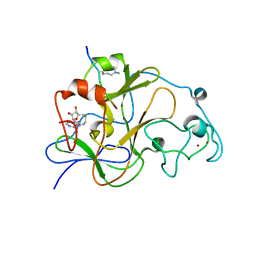 | | GLP (G9a-like protein) SET domain in complex with Dnmt3aK44me2 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone-lysine N-methyltransferase EHMT1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2011-07-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
3SW9
 
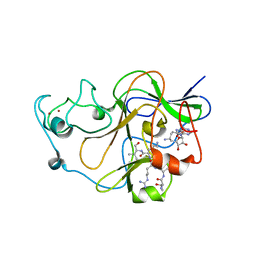 | | GLP (G9a-like protein) SET domain in complex with Dnmt3aK44me0 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone-lysine N-methyltransferase EHMT1, SINEFUNGIN, ... | | Authors: | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2011-07-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
5EH2
 
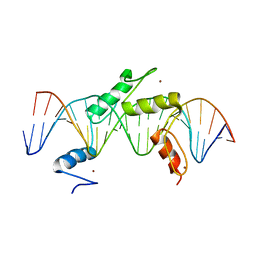 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
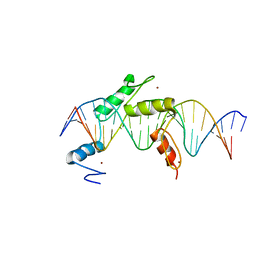 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EGB
 
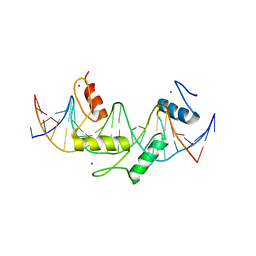 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence II | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
1FJX
 
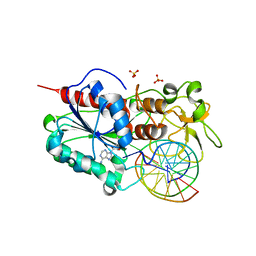 | | STRUCTURE OF TERNARY COMPLEX OF HHAI METHYLTRANSFERASE MUTANT (T250G) IN COMPLEX WITH DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), HHAI DNA METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Vilkaitis, G, Dong, A, Weinhold, E, Cheng, X, Klimasauskas, S. | | Deposit date: | 2000-08-08 | | Release date: | 2000-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase.
J.Biol.Chem., 275, 2000
|
|
1SKM
 
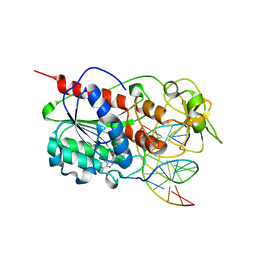 | | HhaI methyltransferase in complex with DNA containing an abasic south carbocyclic sugar at its target site | | Descriptor: | 5'-D(*T*GP*TP*CP*AP*GP*(HCX)P*GP*CP*AP*TP*GP*G)-3', 5'-D(*TP*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', Modification methylase HhaI, ... | | Authors: | Horton, J.R, Ratner, G, Banavali, N, Huang, N, Marquez, V.E, MacKerell, A.D, Cheng, X. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase
Nucleic Acids Res., 32, 2004
|
|
1T43
 
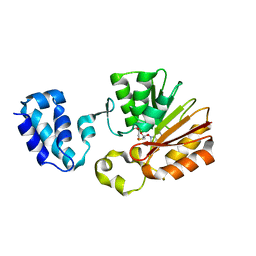 | | Crystal Structure Analysis of E.coli Protein (N5)-Glutamine Methyltransferase (HemK) | | Descriptor: | Protein methyltransferase hemK, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Z, Shipman, L, Zhang, M, Anton, B.P, Roberts, R.J, Cheng, X. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase.
J.Mol.Biol., 340, 2004
|
|
3KV9
 
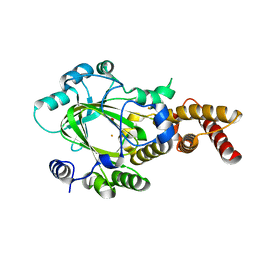 | | Structure of KIAA1718 Jumonji domain | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVB
 
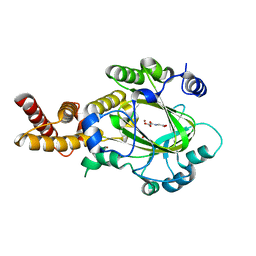 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV6
 
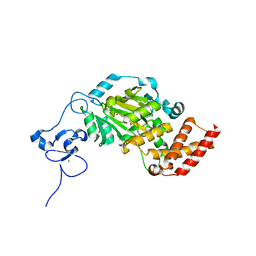 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1YNM
 
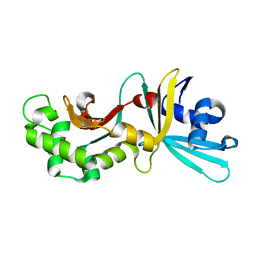 | | Crystal structure of restriction endonuclease HinP1I | | Descriptor: | R.HinP1I restriction endonuclease | | Authors: | Yang, Z, Horton, J.R, Maunus, R, Wilson, G.G, Roberts, R.J, Cheng, X. | | Deposit date: | 2005-01-24 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI
Nucleic Acids Res., 33, 2005
|
|
3KV4
 
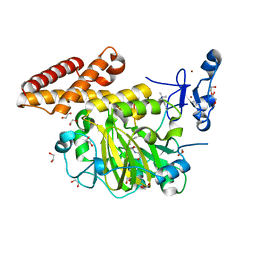 | | Structure of PHF8 in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
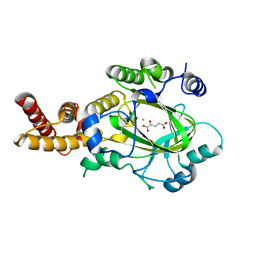 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
