8CXW
 
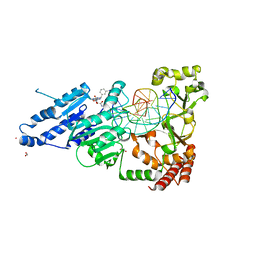 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor piclidenoson (Compound 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY1
 
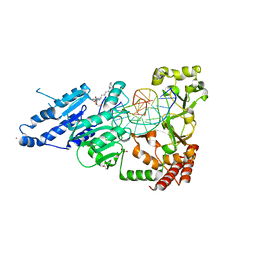 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY4
 
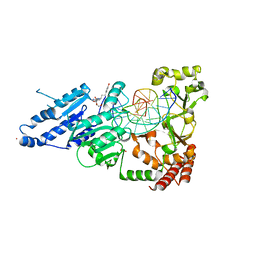 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-[3-(4-hydroxyphenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY0
 
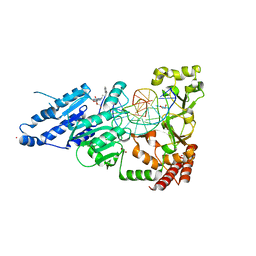 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MC4756 (Compound 178) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(4-phenylbutyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXZ
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(3-Phenylpropyl)adenosine (Compound 14) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(3-phenylpropyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
5J5N
 
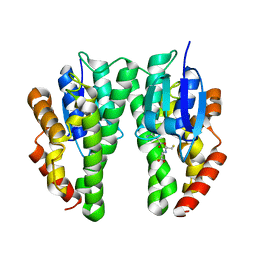 | |
7QXI
 
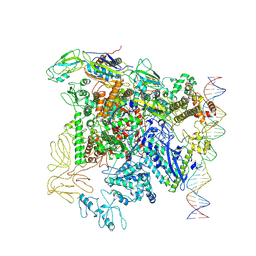 | |
7QWP
 
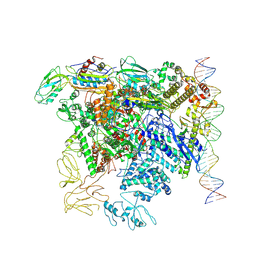 | | CryoEM structure of bacterial transcription close complex (RPc) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ye, F.Z, Zhang, X.D. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms of DNA opening revealed in AAA+ transcription complex structures.
Sci Adv, 8, 2022
|
|
7QV9
 
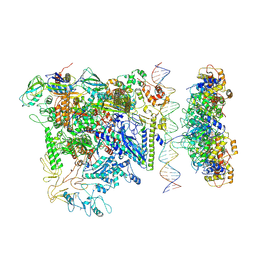 | |
2R7B
 
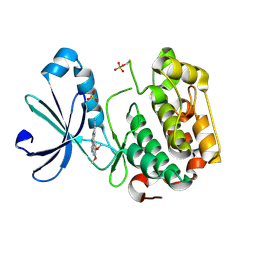 | | Crystal Structure of the Phosphoinositide-dependent Kinase-1 (PDK-1)Catalytic Domain bound to a dibenzonaphthyridine inhibitor | | Descriptor: | 10,11-dimethoxy-4-methyldibenzo[c,f]-2,7-naphthyridine-3,6-diamine, 3-phosphoinositide-dependent protein kinase 1, SULFATE ION | | Authors: | Olland, A.M. | | Deposit date: | 2007-09-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Dibenzo[c,f][2,7]naphthyridines as Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 Inhibitors.
J.Med.Chem., 50, 2007
|
|
8WQW
 
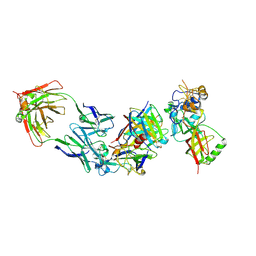 | | Cryo-EM structure of bsAb3 Fab-Gn-Gc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein C, Glycoprotein N, ... | | Authors: | Wu, Y, Sun, J.Q. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5VEB
 
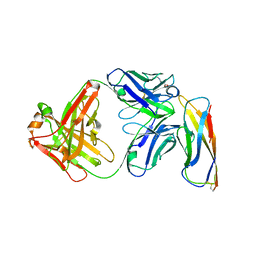 | | Crystal structure of a Fab binding to extracellular domain 5 of Cadherin-6 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Cadherin-6, anti-CDH6 Fab heavy chain, ... | | Authors: | Zhu, X, Bialucha, C.U, London, A, Clark, K, Hu, T. | | Deposit date: | 2017-04-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Optimization of HKT288, a Cadherin-6-Targeting ADC for the Treatment of Ovarian and Renal Cancers.
Cancer Discov, 7, 2017
|
|
5J4U
 
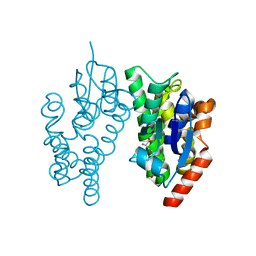 | |
2K8H
 
 | |
4RFY
 
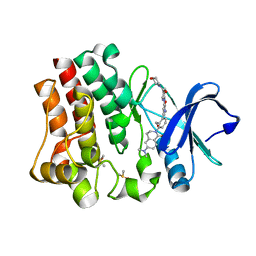 | | Crystal structure of BTK kinase domain complexed with 6-(dimethylamino)-2-[2-(hydroxymethyl)-3-[1-methyl-5-[[5-(morpholine-4-carbonyl)-2-pyridyl]amino]-6-oxo-3-pyridyl]phenyl]-3,4-dihydroisoquinolin-1-one | | Descriptor: | 6-(dimethylamino)-2-[2-(hydroxymethyl)-3-(1-methyl-5-{[5-(morpholin-4-ylcarbonyl)pyridin-2-yl]amino}-6-oxo-1,6-dihydropyridin-3-yl)phenyl]-3,4-dihydroisoquinolin-1(2H)-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Finding the perfect spot for fluorine: Improving potency up to 40-fold during a rational fluorine scan of a Bruton's Tyrosine Kinase (BTK) inhibitor scaffold.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4RG0
 
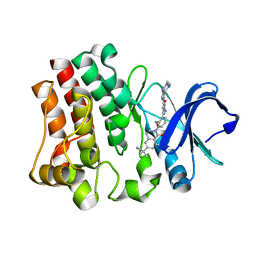 | | Crystal structure of BTK kinase domain complexed with 2-[8-fluoro-2-[2-(hydroxymethyl)-3-[1-methyl-5-[[5-(4-methylpiperazin-1-yl)-2-pyridyl]amino]-6-oxo-3-pyridyl]phenyl]-1-oxo-3,4-dihydroisoquinolin-6-yl]-2-methyl-propanenitrile | | Descriptor: | 2-{8-fluoro-2-[2-(hydroxymethyl)-3-(1-methyl-5-{[5-(4-methylpiperazin-1-yl)pyridin-2-yl]amino}-6-oxo-1,6-dihydropyridin-3-yl)phenyl]-1-oxo-1,2,3,4-tetrahydroisoquinolin-6-yl}-2-methylpropanenitrile, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Finding the perfect spot for fluorine: Improving potency up to 40-fold during a rational fluorine scan of a Bruton's Tyrosine Kinase (BTK) inhibitor scaffold.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7V6G
 
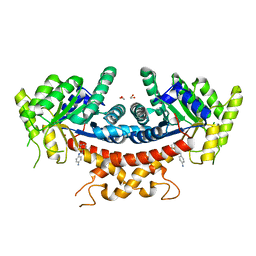 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | Authors: | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
6GH5
 
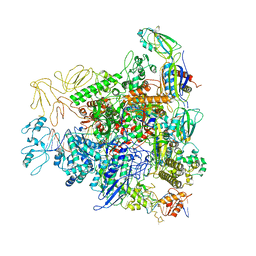 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme transcription open complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6GFW
 
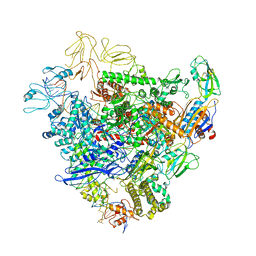 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
4RKY
 
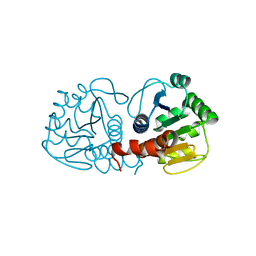 | | Crystal structure of DJ-1 isoform X1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
6GH6
 
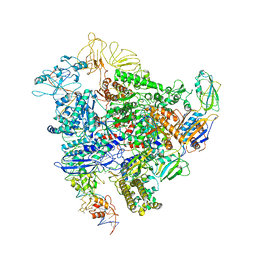 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme intermediate partially loaded complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
4PTC
 
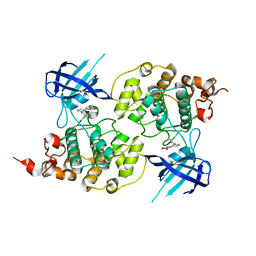 | | Structure of a carboxamide compound (3) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-OXO-4H-1LAMBDA~4~,3-THIAZOLE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-[2-(cyclopropylcarbonylamino)pyridin-4-yl]-4-methoxy-1,3-thiazole-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4RFZ
 
 | |
8TXG
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 8 | | Descriptor: | (4M)-4-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-7-fluoro-1,3-benzothiazol-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8TXH
 
 | | Crystal structure of KRAS G12D in complex with GDP and compound 14 | | Descriptor: | (4P)-2-amino-4-{4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}-6-(trifluoromethyl)quinazolin-7-yl}-7-fluoro-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
