2WU7
 
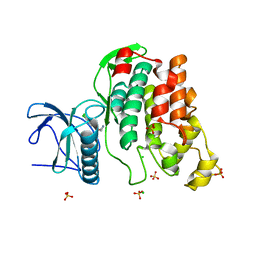 | | Crystal Structure of the Human CLK3 in complex with V25 | | Descriptor: | CHLORIDE ION, DUAL SPECIFICITY PROTEIN KINASE CLK3, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Phillips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Bracher, F, Huber, K, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
2WU6
 
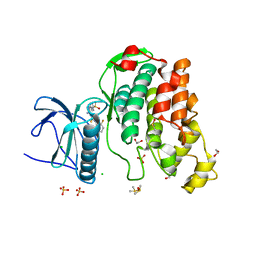 | | Crystal Structure of the Human CLK3 in complex with DKI | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CHLORIDE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Philips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
4CUG
 
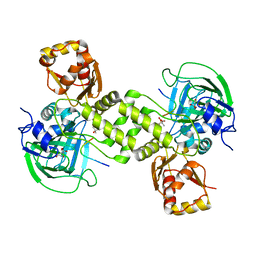 | | Rhodothermus marinus YCFD-like ribosomal protein L16 Arginyl hydroxylase in complex substrate fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S RIBOSOMAL PROTEIN L16, CUPIN 4 FAMILY PROTEIN, ... | | Authors: | McDonough, M.A, Sekirnik, R, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2V1W
 
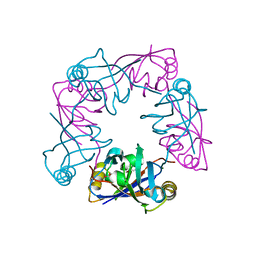 | | Crystal structure of human LIM protein RIL (PDLIM4) PDZ domain bound to the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PDZ AND LIM DOMAIN PROTEIN 4, ... | | Authors: | Soundararajan, M, Shrestha, L, Pike, A.C.W, Salah, E, Burgess-Brown, N, Elkins, J, Umeano, C, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Interactions in Pdz Domain Crystal Structures Help Explain Binding Mechanisms.
Protein Sci., 19, 2010
|
|
3U23
 
 | | Atomic resolution crystal structure of the 2nd SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide from human RIN3 | | Descriptor: | 1,2-ETHANEDIOL, CD2-associated protein, Ras and Rab interactor 3 | | Authors: | Simister, P.C, Rouka, E, Janning, M, Muniz, J.R.C, Kirsch, K.H, Knapp, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Krojer, T, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
2JKV
 
 | | Structure of human Phosphogluconate Dehydrogenase in complex with NADPH at 2.53A | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING, CHLORIDE ION, ... | | Authors: | Pilka, E.S, Kavanagh, K.L, von Delft, F, Muniz, J.R.C, Murray, J, Picaud, S, Guo, K, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2008-09-01 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Structure of Human Phosphogluconate Dehydrogenase in Complex with Nadph at 2.53A
To be Published
|
|
5FLW
 
 | | Crystal structure of putative exo-beta-1,3-galactanase from Bifidobacterium bifidum s17 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EXO-BETA-1,3-GALACTANASE | | Authors: | Godoy, A.S, de Lima, M.Z.T, Ramia, M.P, Camilo, C.M, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of a putative exo-beta-1,3-galactanase from Bifidobacterium bifidum S17.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5OWR
 
 | | Human STK10 bound to dasatinib | | Descriptor: | CALCIUM ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Serine/threonine-protein kinase 10 | | Authors: | Szklarz, M, Muniz, J.R.C, Vollmar, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2017-09-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human STK10 bound to dasatinib
To Be Published
|
|
4LP8
 
 | | A Novel Open-State Crystal Structure of the Prokaryotic Inward Rectifier KirBac3.1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Zubcevic, L, Bavro, V.N, Muniz, J.R.C, Schmidt, M.R, Wang, S, De Zorzi, R, Venien-Bryan, C, Sansom, M.S.P, Nichols, C.G, Tucker, S.J. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Control of KirBac3.1 Potassium Channel Gating at the Interface between Cytoplasmic Domains.
J.Biol.Chem., 289, 2014
|
|
4DYO
 
 | | Crystal Structure of Human Aspartyl Aminopeptidase (DNPEP) in complex with Aspartic acid Hydroxamate | | Descriptor: | Aspartyl aminopeptidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chaikuad, A, Pilka, E, Vollmar, M, Krojer, T, Muniz, J.R.C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human aspartyl aminopeptidase complexed with substrate analogue: insight into catalytic mechanism, substrate specificity and M18 peptidase family.
Bmc Struct.Biol., 12, 2012
|
|
3MY0
 
 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | Authors: | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
3KHU
 
 | | Crystal structure of human UDP-glucose dehydrogenase Glu161Gln, in complex with thiohemiacetal intermediate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, UDP-glucose 6-dehydrogenase, ... | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Guo, K, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Evidence That Catalytic Reaction of Human UDP-glucose 6-Dehydrogenase Involves Covalent Thiohemiacetal and Thioester Enzyme Intermediates.
J.Biol.Chem., 287, 2012
|
|
3H9R
 
 | | Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin | | Descriptor: | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Activin receptor type-1, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Chaikuad, A, Alfano, I, Shrestha, B, Muniz, J.R.C, Petrie, K, Fedorov, O, Phillips, C, Bishop, S, Mahajan, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Bone Morphogenetic Protein Receptor ALK2 and Implications for Fibrodysplasia Ossificans Progressiva.
J.Biol.Chem., 287, 2012
|
|
3II7
 
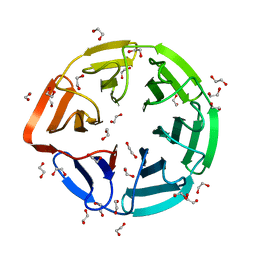 | | Crystal structure of the kelch domain of human KLHL7 | | Descriptor: | 1,2-ETHANEDIOL, Kelch-like protein 7 | | Authors: | Chaikuad, A, Thangaratnarajah, C, Cooper, C.D.O, Ugochukwu, E, Muniz, J.R.C, Krojer, T, Sethi, R, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases.
J.Biol.Chem., 288, 2013
|
|
3ITK
 
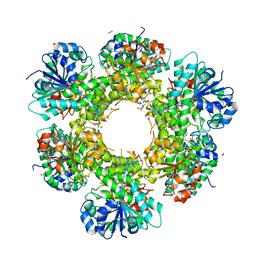 | | Crystal structure of human UDP-glucose dehydrogenase Thr131Ala, apo form. | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, UDP-glucose 6-dehydrogenase | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
4CCL
 
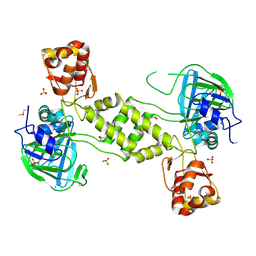 | | X-Ray structure of E. coli ycfD | | Descriptor: | 50S RIBOSOMAL PROTEIN L16 ARGININE HYDROXYLASE, MANGANESE (II) ION, SULFATE ION | | Authors: | McDonough, M.A, Ho, C.H, Kershaw, N.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4BXF
 
 | |
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCM
 
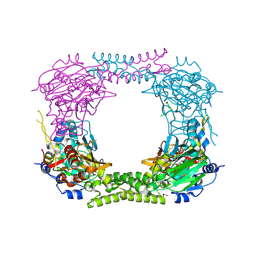 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-1) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCO
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 S373C) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G214C) peptide fragment (complex-3) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCN
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 L299C/C300S) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-2) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCK
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II) and N-oxalylglycine (NOG) | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4BU2
 
 | | 60S ribosomal protein L27A histidine hydroxylase (MINA53) in complex with Ni(II) and 2-oxoglutarate (2OG) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE MINA, ... | | Authors: | Chowdhury, R, Clifton, I.J, McDonough, M.A, Ng, S.S, Pilka, E, Oppermann, U, Schofield, C.J. | | Deposit date: | 2013-06-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CSW
 
 | | Rhodothermus marinus YCFD-like ribosomal protein L16 Arginyl hydroxylase | | Descriptor: | CITRIC ACID, CUPIN 4 FAMILY PROTEIN, GLYCEROL, ... | | Authors: | McDonough, M.A, Sekirnik, R, Schofield, C.J. | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2YOK
 
 | | Cellobiohydrolase I Cel7A from Trichoderma harzianum at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Textor, L.C, Colussi, F, Serpa, V, Squina, F.M, Pereira Jr, N, Polikarpov, I. | | Deposit date: | 2012-10-25 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Joint X-Ray Crystallographic and Molecular Dynamics Study of Cellobiohydrolase I from Trichoderma Harzianum: Deciphering the Structural Features of Cellobiohydrolase Catalytic Activity.
FEBS J., 280, 2013
|
|
