5PGX
 
 | |
5PGY
 
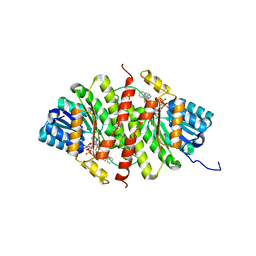 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
7KDT
 
 | |
5PGZ
 
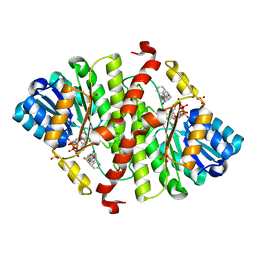 | | CRYSTAL STRUCTURE OF MURINE 11BETA- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7EPZ
 
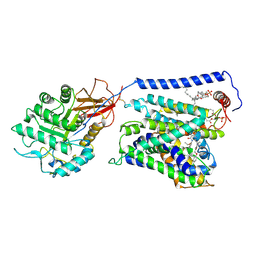 | | Overall structure of Erastin-bound xCT-4F2hc complex | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-[(1S)-1-[4-[2-(4-chloranylphenoxy)ethanoyl]piperazin-1-yl]ethyl]-3-(2-ethoxyphenyl)quinazolin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis.
Cell Res., 32, 2022
|
|
5JWB
 
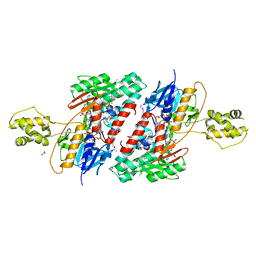 | | Structure of NDH2 from plasmodium falciparum in complex with NADH | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yu, Y, Li, X.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
6E8N
 
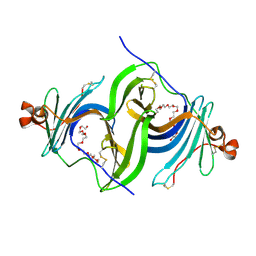 | | Crystal structure of glycosylated human EPDR1 | | Descriptor: | Mammalian ependymin-related protein 1, NONAETHYLENE GLYCOL | | Authors: | Wei, Y, Prive, G.G. | | Deposit date: | 2018-07-30 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of human lysosomal EPDR1 reveal homology with the superfamily of bacterial lipoprotein transporters.
Commun Biol, 2, 2019
|
|
7RN5
 
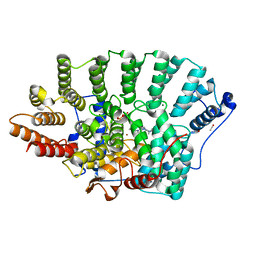 | |
7RNI
 
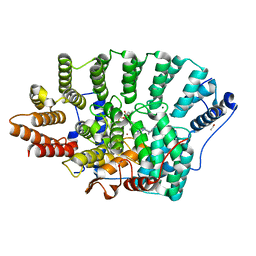 | |
7EXC
 
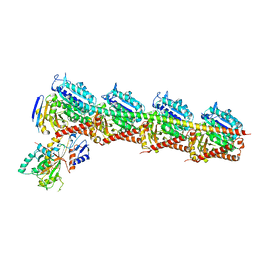 | | Crystal structure of T2R-TTL-1129A2 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2021-05-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-Based Design and Synthesis of N-Substituted 3-Amino-beta-Carboline Derivatives as Potent alpha beta-Tubulin Degradation Agents
J.Med.Chem., 65, 2022
|
|
5X72
 
 | | The crystal Structure PDE delta in complex with (rac)-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, (2S)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
8I5N
 
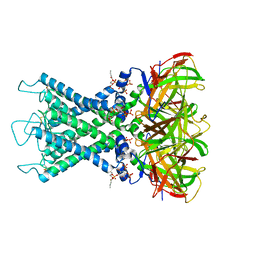 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
8I5M
 
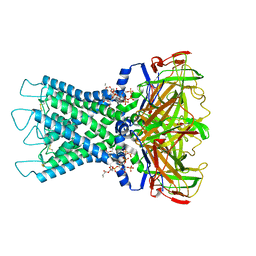 | | Rat Kir4.1 in complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
9LDS
 
 | |
6FBA
 
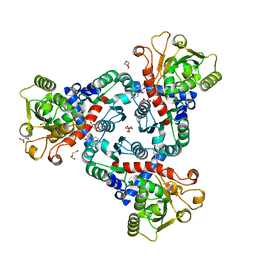 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with bound inhibitor 2,3-naphthalenediol | | Descriptor: | Aspartate transcarbamoylase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.A, Wang, C, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a non-competitive inhibitor of Plasmodium falciparum aspartate transcarbamoylase.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
8HOK
 
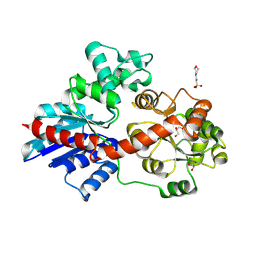 | | crystal structure of UGT71AP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, UGT71AP2 | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
8HOJ
 
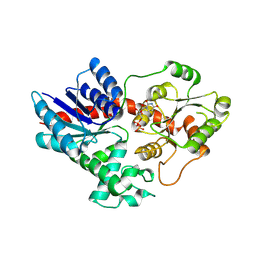 | | Crystal structure of UGT71AP2 in complex with UDP | | Descriptor: | UGT71AP2, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
9LON
 
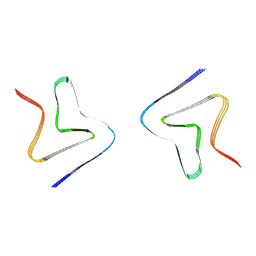 | | Cryo-EM structure of alpha-synuclein P8 fibril | | Descriptor: | Alpha-synuclein | | Authors: | Xia, W.C, Liu, C. | | Deposit date: | 2025-01-23 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fibril fuzzy coat is important for alpha-synuclein pathological transmission activity.
Neuron, 113, 2025
|
|
8I0D
 
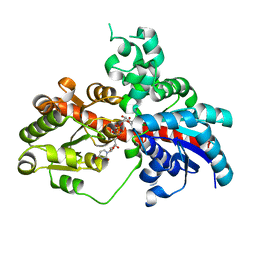 | | Sb3GT1 375S/Q377H mutant complex with UDP-Glc | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
5JWC
 
 | | Structure of NDH2 from plasmodium falciparum in complex with RYL-552 | | Descriptor: | 5-fluoro-3-methyl-2-{4-[4-(trifluoromethoxy)benzyl]phenyl}quinolin-4(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
