5W8O
 
 | | Homoserine transacetylase MetX from Mycobacterium hassiacum | | Descriptor: | CALCIUM ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Reed, R.W, Rodriguez, E.S, Li, J, Korotkov, K.V. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural analysis of mycobacterial homoserine transacetylases central to methionine biosynthesis reveals druggable active site.
Sci Rep, 9, 2019
|
|
7E6R
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.6 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-23 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6L
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.0 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78037143 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6M
 
 | | Crystal structure of Human coronavirus NL63 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83445024 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6N
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.2 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8413 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8HHV
 
 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
7XB3
 
 | |
7DYS
 
 | |
1SZJ
 
 | |
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4HU8
 
 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
7YL2
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4J0U
 
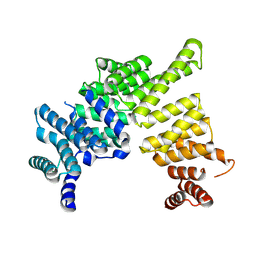 | | Crystal structure of IFIT5/ISG58 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5 | | Authors: | Liu, Y, Liang, H, Feng, F, Yuan, L, Wang, Y.E, Crowley, C, Lv, Z, Li, J, Zeng, S, Cheng, G. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Crystal Structure of IFIT5
To be Published
|
|
7E27
 
 | | Structure of PfFNT in complex with MMV007839 | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7E26
 
 | | Structure of PfFNT in apo state | | Descriptor: | Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7FAI
 
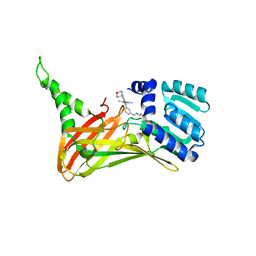 | | CARM1 bound with compound 9 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-(oxan-4-ylamino)pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09749269 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
7FAJ
 
 | | CARM1 bound with compound 43 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-phenylazanyl-pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2450726 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
4HY6
 
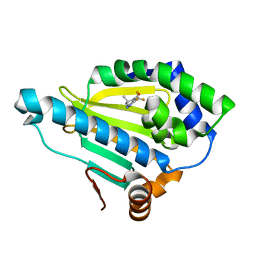 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1 | | Descriptor: | 6,6-dimethyl-3-(trifluoromethyl)-1,5,6,7-tetrahydro-4H-indazol-4-one, Heat shock protein HSP 90-alpha | | Authors: | Yang, M, Li, J, He, J.H. | | Deposit date: | 2012-11-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1
to be published
|
|
7C2Y
 
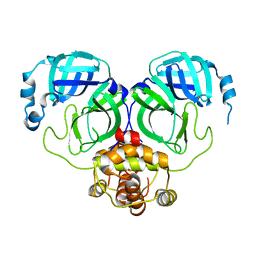 | | The crystal structure of COVID-2019 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | COVID-2019 main protease in the apo state
To Be Published
|
|
7CCD
 
 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*GP*AS*AP*CP*GP*TP*G)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Li, J, Liu, G, Zhao, G, Wang, Y, Hu, W, Deng, Z, Gan, J, Zhao, Y, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7DR8
 
 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7DR9
 
 | |
7DRA
 
 | |
5Y4D
 
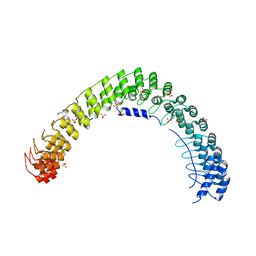 | | Crystal Structure of AnkB Ankyrin Repeats in Complex with AnkR/AnkB Chimeric Autoinhibition Segment | | Descriptor: | Ankyrin-1,Ankyrin-2,Ankyrin-2, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
