7NSC
 
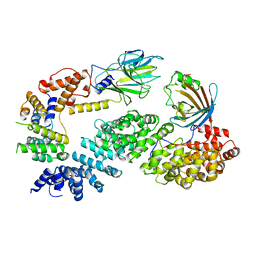 | | Substrate receptor scaffolding module of human CTLH E3 ubiquitin ligase | | Descriptor: | Glucose-induced degradation protein 4 homolog, Glucose-induced degradation protein 8 homolog, Isoform 2 of Armadillo repeat-containing protein 8, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NS4
 
 | | Catalytic module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, ZINC ION | | Authors: | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
5UDH
 
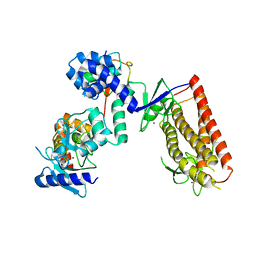 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
5V89
 
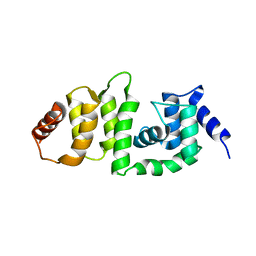 | | Structure of DCN4 PONY domain bound to CUL1 WHB | | Descriptor: | Cullin-1, DCN1-like protein 4 | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V83
 
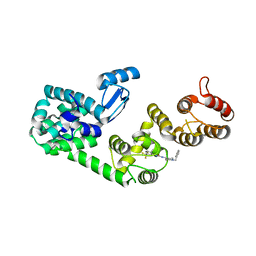 | | Structure of DCN1 bound to NAcM-HIT | | Descriptor: | Lysozyme,DCN1-like protein 1 chimera, N-(1-benzylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
4YII
 
 | |
8CDK
 
 | |
8CDJ
 
 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
8C07
 
 | |
8C06
 
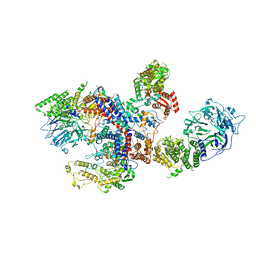 | |
8CAF
 
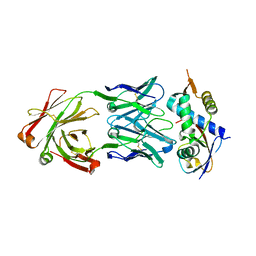 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | Descriptor: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
4KBL
 
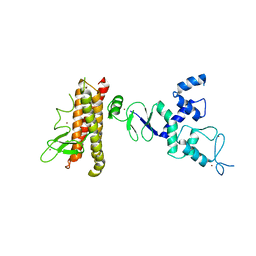 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
4KC9
 
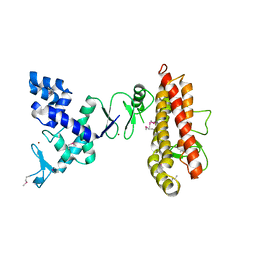 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
4LCD
 
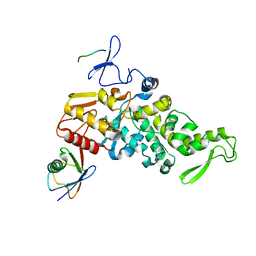 | |
3DBL
 
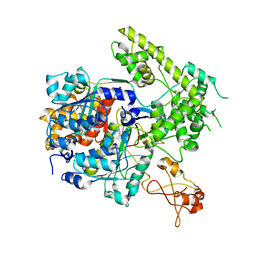 | |
3DBH
 
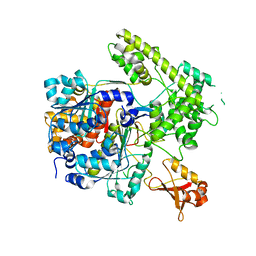 | |
3DPL
 
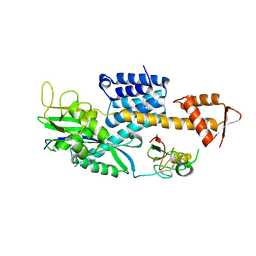 | |
3DBR
 
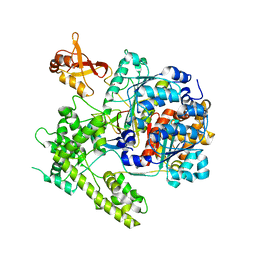 | |
3DQV
 
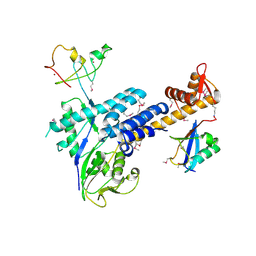 | | Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation | | Descriptor: | Cullin-5, NEDD8, Rbx1, ... | | Authors: | Duda, D.M, Borg, L.A, Scott, D.C, Hunt, H.W, Hammel, M, Schulman, B.A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Cell(Cambridge,Mass.), 134, 2008
|
|
2M6N
 
 | | 3D solution structure of EMI1 (Early Mitotic Inhibitor 1) | | Descriptor: | F-box only protein 5, ZINC ION | | Authors: | Frye, J.J, Brown, N.G, Petzold, G, Watson, E.R, Royappa, G.R, Nourse, A, Jarvis, M, Kriwacki, R.W, Peters, J, Stark, H, Schulman, B.A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2NVU
 
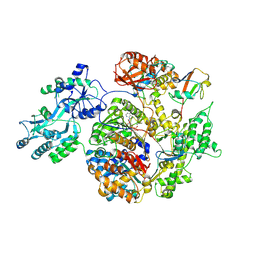 | | Structure of APPBP1-UBA3~NEDD8-NEDD8-MgATP-Ubc12(C111A), a trapped ubiquitin-like protein activation complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maltose binding protein/NEDD8-activating enzyme E1 catalytic subunit chimera, ... | | Authors: | Huang, D.T, Hunt, H.W, Zhuang, M, Ohi, M.D, Holton, J.M, Schulman, B.A. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity.
Nature, 445, 2007
|
|
4P5O
 
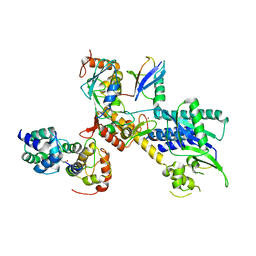 | |
4O1V
 
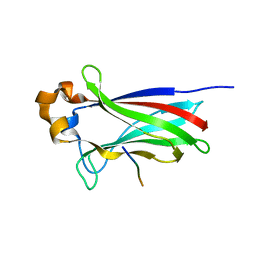 | | SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, Speckle-type POZ protein | | Authors: | Calabrese, M.F, Watson, E.R, Schulman, B.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SPOP Promotes Tumorigenesis by Acting as a Key Regulatory Hub in Kidney Cancer.
Cancer Cell, 25, 2014
|
|
4R2Y
 
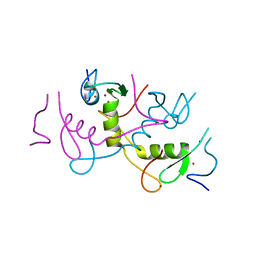 | | Crystal structure of APC11 RING domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissmann, F, Jarvis, M.A, Vanderlinden, R, Grace, C.R.R, Frye, J.J, Dube, P, Qiao, R, Petzold, G, Cho, S.E, Alsharif, O, Bao, J, Zheng, J, Nourse, A, Kurinov, I, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
4RG7
 
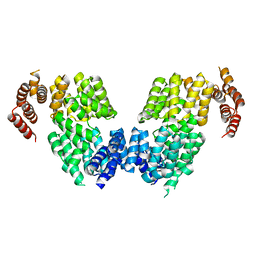 | | Crystal structure of APC3 | | Descriptor: | Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
