8XLO
 
 | |
8GVJ
 
 | | Crystal structure of cMET kinase domain bound by D6808 | | Descriptor: | (1^4Z,5^2E)-6^3-(trifluoromethyl)-5^1,5^6-dihydro-1^1H-8-aza-2(3,6)-quinolina-5(1,3)-pyridazina-1(4,1)-pyrazola-6(1,4)-benzenacyclododecaphane-5^6,7-dione, Hepatocyte growth factor receptor | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2022-09-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wei, H, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
7D42
 
 | |
7DGD
 
 | | apo state of class C GPCR | | Descriptor: | Metabotropic glutamate receptor 1 | | Authors: | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | Deposit date: | 2020-11-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
7DGE
 
 | | intermediate state of class C GPCR | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 1, nanobody | | Authors: | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | Deposit date: | 2020-11-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
7Y4U
 
 | | Crystal structure of cMET kinase domain bound by compound 9Y | | Descriptor: | Hepatocyte growth factor receptor, ~{N}-methyl-4-[1-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-oxidanylidene-pyridazin-3-yl]-2-(trifluoromethyl)benzamide | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7Y4T
 
 | | Crystal structure of cMET kinase domain bound by compound 9I | | Descriptor: | 2-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-(3-nitrophenyl)pyridazin-3-one, Hepatocyte growth factor receptor | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7VUE
 
 | | Structural insight of the molecular mechanism of cilofexor bound to FXR | | Descriptor: | 2-[3-[4-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-2-chloranyl-phenyl]-3-oxidanyl-azetidin-1-yl]pyridine-4-carboxylic acid, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Jiang, L, Chen, Y.C. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural insight into the molecular mechanism of cilofexor binding to the farnesoid X receptor.
Biochem.Biophys.Res.Commun., 595, 2022
|
|
7WNQ
 
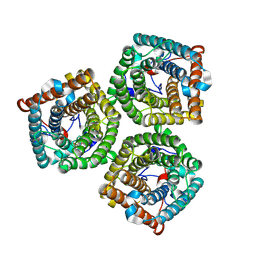 | | Cryo-EM structure of AtSLAC1 S59A mutant | | Descriptor: | Guard cell S-type anion channel SLAC1 | | Authors: | Sun, L, Liu, X, Li, Y. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the Arabidopsis guard cell anion channel SLAC1 suggests activation mechanism by phosphorylation.
Nat Commun, 13, 2022
|
|
7WF5
 
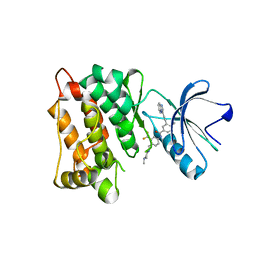 | | c-Src in complex with ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Duan, Y, Dai, S, Chen, X, Chen, Y. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural study of ponatinib in inhibiting SRC kinase.
Biochem.Biophys.Res.Commun., 598, 2022
|
|
7VNI
 
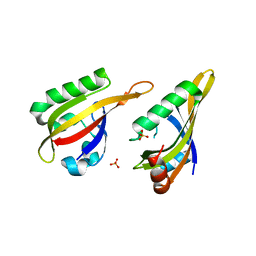 | | AHR-ARNT PAS-B heterodimer | | Descriptor: | Ahr homolog spineless, Aryl hydrocarbon receptor nuclear translocator, SULFATE ION | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNH
 
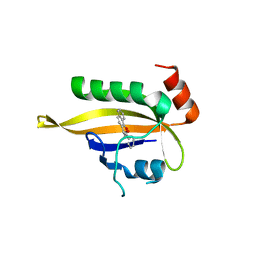 | |
7VNA
 
 | | drosophlia AHR PAS-B domain | | Descriptor: | Ahr homolog spineless | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-10 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
8JOT
 
 | | Crystal structure of CSF-1R kinase domain with sulfatinib | | Descriptor: | GLYCEROL, Macrophage colony-stimulating factor 1 receptor, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
8JMZ
 
 | | FGFR1 kinase domain with sulfatinib | | Descriptor: | Fibroblast growth factor receptor 1, GLYCEROL, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
8IFO
 
 | | Crystal structure of estrogen related receptor-gamma DNA binding domain complexed with Pla2g12b promoter | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*CP*AP*AP*AP*GP*GP*TP*GP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*CP*AP*CP*CP*TP*TP*TP*GP*TP*CP*CP*TP*C)-3'), Estrogen-related receptor gamma, ... | | Authors: | Xu, T, Zhen, X, Liu, J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ERR gamma-DBD undergoes dimerization and conformational rearrangement upon binding to the downstream site of the DR1 element.
Biochem.Biophys.Res.Commun., 656, 2023
|
|
