7D5O
 
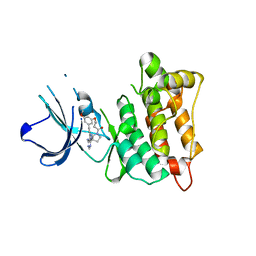 | | C-Src in complex with TAS-120 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2020-09-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7X4W
 
 | |
7X4X
 
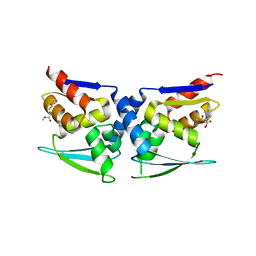
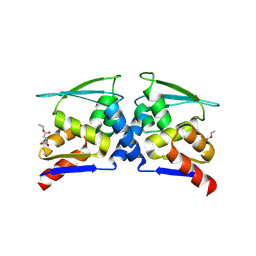 | | BTB domain of KEAP1 in complex with MEF | | Descriptor: | 4-ethoxy-4-oxobutanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Qu, L.Z. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Characterization of the modification of Kelch-like ECH-associated protein 1 by different fumarates.
Biochem.Biophys.Res.Commun., 605, 2022
|
|
7Y4T
 
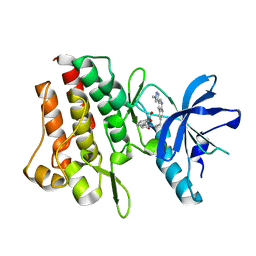 | | Crystal structure of cMET kinase domain bound by compound 9I | | Descriptor: | 2-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-(3-nitrophenyl)pyridazin-3-one, Hepatocyte growth factor receptor | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7Y4U
 
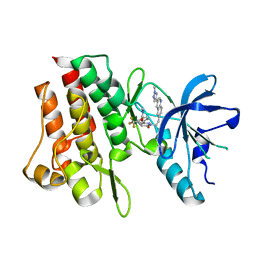 | | Crystal structure of cMET kinase domain bound by compound 9Y | | Descriptor: | Hepatocyte growth factor receptor, ~{N}-methyl-4-[1-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-oxidanylidene-pyridazin-3-yl]-2-(trifluoromethyl)benzamide | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7F3M
 
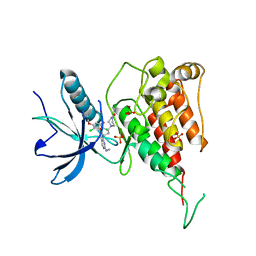 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
8GVJ
 
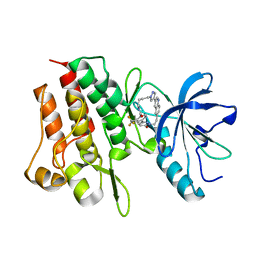 | | Crystal structure of cMET kinase domain bound by D6808 | | Descriptor: | (1^4Z,5^2E)-6^3-(trifluoromethyl)-5^1,5^6-dihydro-1^1H-8-aza-2(3,6)-quinolina-5(1,3)-pyridazina-1(4,1)-pyrazola-6(1,4)-benzenacyclododecaphane-5^6,7-dione, Hepatocyte growth factor receptor | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2022-09-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7V29
 
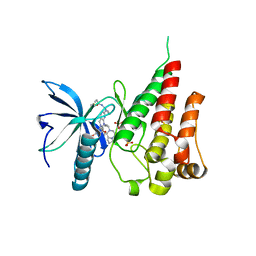 | | Crystal structure of FGFR4 with a dual-warhead covalent inhhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[3-(3,5-dimethoxyphenyl)-2-oxidanylidene-1-[3-(4-propanoylpiperazin-1-yl)propyl]-4H-pyrimido[4,5-d]pyrimidin-7-yl]amino]phenyl]propanamide, SULFATE ION | | Authors: | Chen, X.J, Jiang, L.Y, Dai, S.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-08-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Structure-based design of a dual-warhead covalent inhibitor of FGFR4.
Commun Chem, 5, 2022
|
|
7D57
 
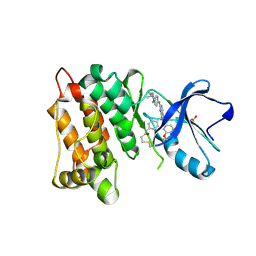 | | C-Src in complex with FIIN-2 | | Descriptor: | N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7WCL
 
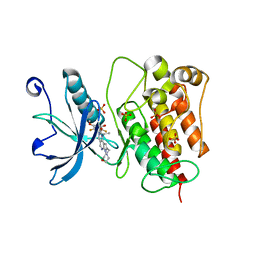 | | Crystal structure of FGFR1 kinase domain with Pemigatinib | | Descriptor: | 11-[2,6-bis(fluoranyl)-3,5-dimethoxy-phenyl]-13-ethyl-4-(morpholin-4-ylmethyl)-5,7,11,13-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),2(6),3,7-tetraen-12-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Lin, Q.M, Jiang, L.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Characterization of the cholangiocarcinoma drug pemigatinib against FGFR gatekeeper mutants.
Commun Chem, 5, 2022
|
|
