6L4R
 
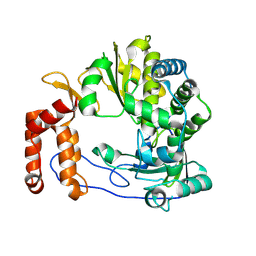 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
7YE1
 
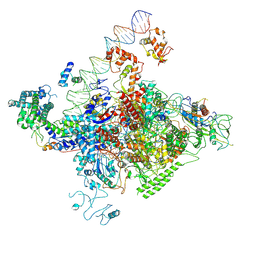 | | The cryo-EM structure of C. crescentus GcrA-TACup | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (57-MER)-non template, DNA (57-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7YE2
 
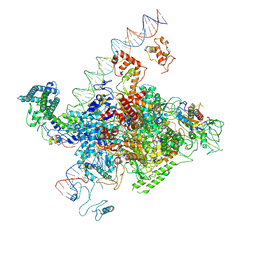 | | The cryo-EM structure of C. crescentus GcrA-TACdown | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (90-MER)-non template, DNA (90-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7SQ7
 
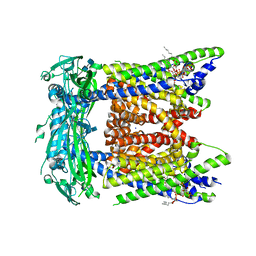 | | Cryo-EM structure of mouse PI(3,5)P2-bound TRPML1 channel at 2.41 Angstrom resolution | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ8
 
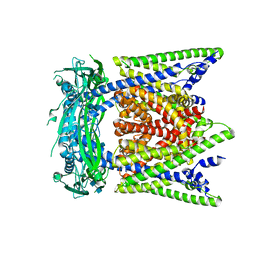 | | Cryo-EM structure of mouse apo TRPML1 channel at 2.598 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.598 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ9
 
 | | Cryo-EM structure of mouse temsirolimus/PI(3,5)P2-bound TRPML1 channel at 2.11 Angstrom resolution | | Descriptor: | (1R,2R,4S)-4-{(2R)-2-[(3S,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30S,34aS)-9,27-dihydroxy-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-1,5,11,28,29-pentaoxo-1,4,5,6,9,10,11,12,13,14,21,22,23,24,25,26,27,28,29,31,32,33,34,34a-tetracosahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontin-3-yl]propyl}-2-methoxycyclohexyl 3-hydroxy-2-(hydroxymethyl)-2-methylpropanoate, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ6
 
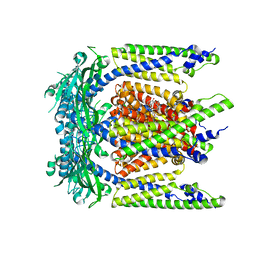 | | Cryo-EM structure of mouse agonist ML-SA1-bound TRPML1 channel at 2.32 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6K3B
 
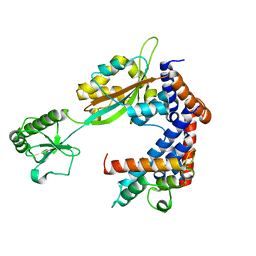 | | Crystal structure of Lpg2147-Lpg2149 complex | | Descriptor: | Lpg2147, Uncharacterized protein | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
6KFP
 
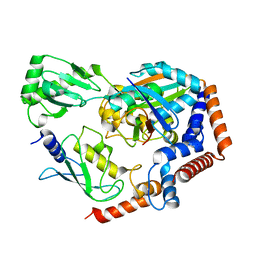 | | Crystal structure of MavC ternary complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
7N7P
 
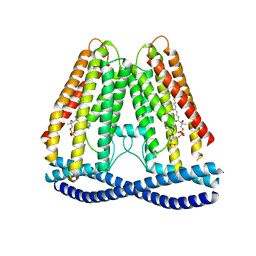 | | Cryo-EM structure of human TMEM120A | | Descriptor: | COENZYME A, Ion channel TACAN | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMEM120A is a coenzyme A-binding membrane protein with structural similarities to ELOVL fatty acid elongase.
Elife, 10, 2021
|
|
6VUA
 
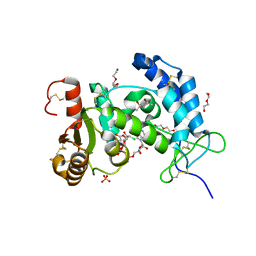 | | X-ray structure of human CD38 catalytic domain with 2'-Cl-araNAD+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, Z, Zhang, X.N, Nasertorabi, F, Han, G.W, Stevens, R.C, Zhang, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis of site-specific antibody-drug conjugates by ADP-ribosyl cyclases.
Sci Adv, 6, 2020
|
|
6DJ1
 
 | | Wild-type HIV-1 protease in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6DJ2
 
 | | HIV-1 protease with single mutation L76V in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6L94
 
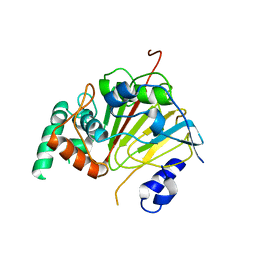 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
6DDE
 
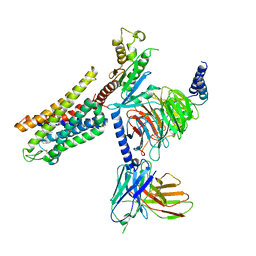 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Zhang, Y, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
6LDI
 
 | |
3JSG
 
 | |
3JTU
 
 | |
3JSF
 
 | |
3L5T
 
 | |
6KKZ
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of perdeuterated GFP at pD 8.5 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
8ISS
 
 | | Cryo-EM structure of wild-type human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG at 3.19 A resolution | | Descriptor: | MAGNESIUM ION, RNA (88-MER), tRNA-splicing endonuclease subunit Sen15, ... | | Authors: | Sun, Y, Zhang, Y, Yuan, L, Han, Y. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex.
Nat Commun, 14, 2023
|
|
3L5S
 
 | |
3L5V
 
 | |
8E3S
 
 | |
