2R9M
 
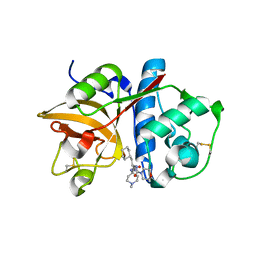 | | Cathepsin S complexed with Compound 15 | | Descriptor: | Cathepsin S, N-[(1S)-2-[(4-cyano-1-methylpiperidin-4-yl)amino]-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
To be Published
|
|
2R9O
 
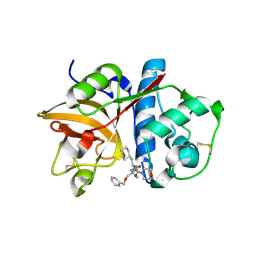 | | Cathepsin S complexed with Compound 8 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(1R)-2-(benzyloxy)-1-cyano-1-methylethyl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
2R9N
 
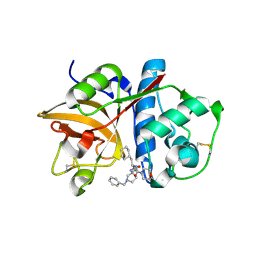 | | Cathepsin S complexed with Compound 26 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(3S)-1-benzyl-3-cyanopyrrolidin-3-yl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
7LJC
 
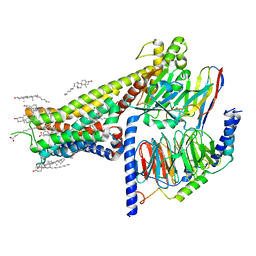 | | Allosteric modulator LY3154207 binding to SKF-81297-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
7LJD
 
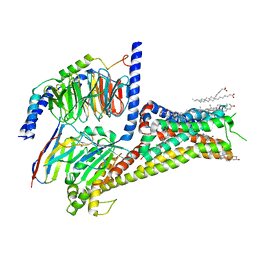 | | Allosteric modulator LY3154207 binding to dopamine-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
4KPY
 
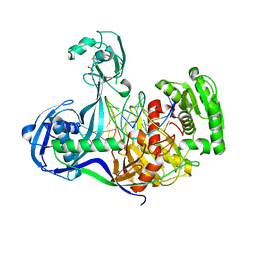 | | DNA binding protein and DNA complex structure | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7XZ4
 
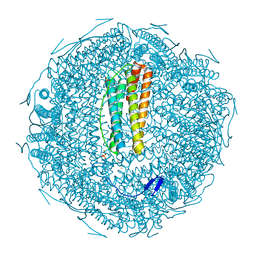 | |
7YDX
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound RI-962 | | Descriptor: | 1-methyl-5-[2-(2-methylpropanoylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-phenylethyl]indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Wu, C, Luo, X, Wang, T, Lei, J, Yang, S. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor.
Nat Commun, 13, 2022
|
|
4BS1
 
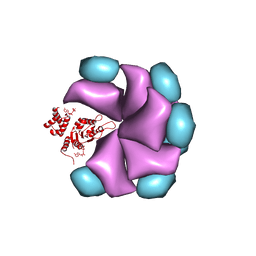 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR (NTRC FAMILY) | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7JVQ
 
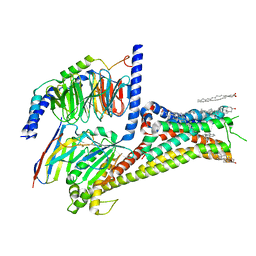 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVP
 
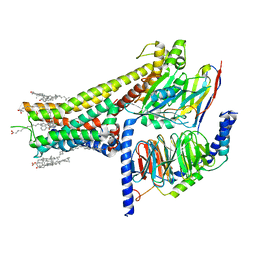 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
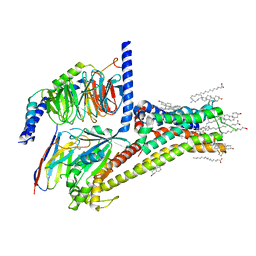 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVR
 
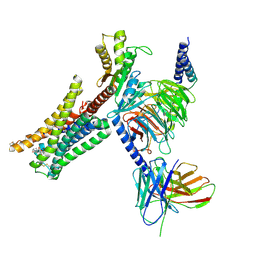 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
1MRJ
 
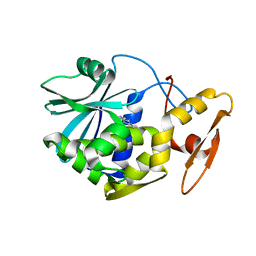 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRG
 
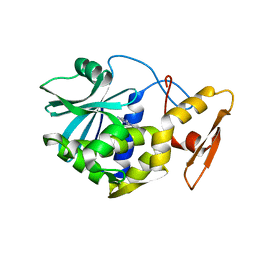 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
4NCA
 
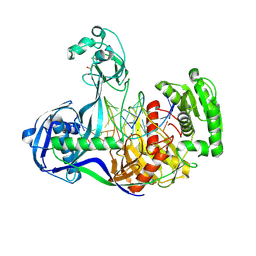 | | Structure of Thermus thermophilus Argonaute bound to guide DNA 19-mer and target DNA in the presence of Mg2+ | | Descriptor: | 5'-D(*AP*CP*AP*AP*CP*C)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BGG
 
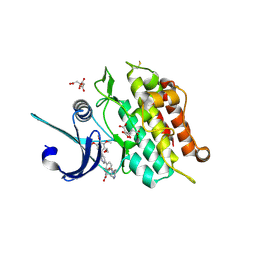 | | Crystal structure of the ACVR1 kinase in complex with LDN-213844 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl}piperazine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Sanvitale, C, Canning, P, Cooper, C, Wang, Y, Mohedas, A.H, Choi, S, Yu, P.B, Cuny, G.D, Nowak, R, Coutandin, D, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure-Activity Relationship of 3,5-Diaryl-2-Aminopyridine Alk2 Inhibitors Reveals Unaltered Binding Affinity for Fibrodysplasia Ossificans Progressiva Causing Mutants.
J.Med.Chem., 57, 2014
|
|
4N47
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 12-mer target DNA | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.823 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BT1
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N41
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | Descriptor: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BT0
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
4H75
 
 | | Crystal structure of human Spindlin1 in complex with a histone H3K4(me3) peptide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Histone H3, ... | | Authors: | Yang, N, Wang, W, Wang, Y, Wang, M, Zhao, Q, Rao, Z, Zhu, B, Xu, R.M. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1MRI
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
4OBY
 
 | | Crystal Structure of E.coli Arginyl-tRNA Synthetase and Ligand Binding Studies Revealed Key Residues in Arginine Recognition | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Bi, K, Zheng, Y, Dong, J, Gao, F, Wang, J, Wang, Y, Gong, W. | | Deposit date: | 2014-01-08 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Crystal structure of E. coli arginyl-tRNA synthetase and ligand binding studies revealed key residues in arginine recognition.
Protein Cell, 5, 2014
|
|
