5D75
 
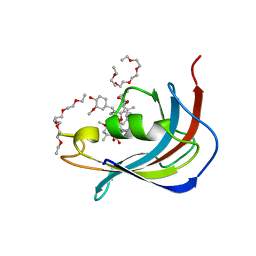 | | Crystal structure of Human FKBD25 in complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Peptidyl-prolyl cis-trans isomerase FKBP3 | | Authors: | Rajan, S, Prakash, A, Yoon, H.S. | | Deposit date: | 2015-08-13 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the FK506 binding domain of human FKBP25 in complex with FK506.
Protein Sci., 25, 2016
|
|
3TP4
 
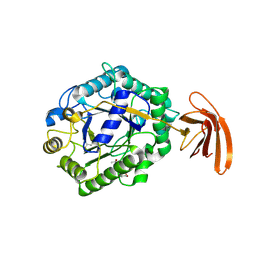 | | Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETK
 
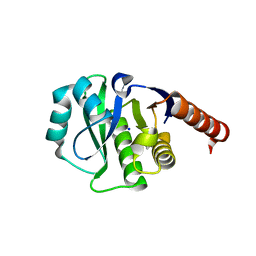 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ESS
 
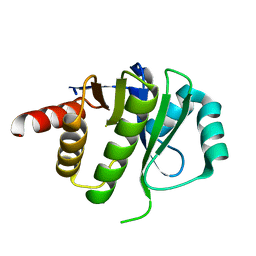 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4MGV
 
 | |
3T03
 
 | | Crystal structure of PPAR gamma ligand binding domain in complex with a novel partial agonist GQ-16 | | Descriptor: | (5Z)-5-(5-bromo-2-methoxybenzylidene)-3-(4-methylbenzyl)-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Rajagopalan, S, Webb, P, Baxter, J.D, Brennan, R.G, Phillips, K.J. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
J.Biol.Chem., 287, 2012
|
|
4KYB
 
 | | Crystal Structure of de novo designed serine hydrolase OSH55.14_E3, Northeast Structural Genomics Consortium Target OR342 | | Descriptor: | Designed Protein OR342, PHOSPHATE ION | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Raja, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR342
To be Published
|
|
8GOD
 
 | | Co-crystal structure of Human Protein-arginine deiminase type-4 (PAD4) with small molecule inhibitor JBI-589 | | Descriptor: | Protein-arginine deiminase type-4, [(3~{R})-3-azanylpiperidin-1-yl]-[2-[1-[(4-fluorophenyl)methyl]indol-2-yl]-3-methyl-imidazo[1,2-a]pyridin-7-yl]methanone | | Authors: | Swaminathan, S, Birudukota, S, Vaithilingam, K, Kandan, S, Asaithambi, K, Kathiresan, N, Gosu, R, Rajagopal, S, Sadhu, N. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Alleviation of arthritis through prevention of neutrophil extracellular traps by an orally available inhibitor of protein arginine deiminase 4.
Sci Rep, 13, 2023
|
|
5YJF
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
5YJI
 
 | | Co-crystal structure of Mouse Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
4EC2
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
5ZQU
 
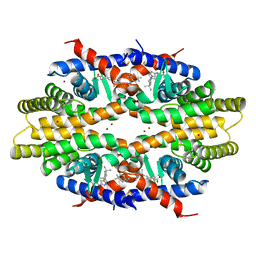 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
1S4S
 
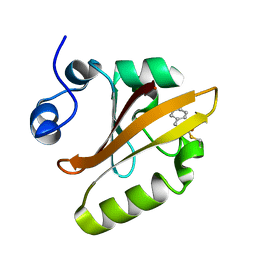 | | Reaction Intermediate in the Photocycle of PYP, intermediate occupied between 100 micro-seconds to 5 milli-seconds | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Schmidt, M, Pahl, R, Srajer, V, Anderson, S, Ren, Z, Ihee, H, Rajagopal, S, Moffat, K. | | Deposit date: | 2004-01-17 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein kinetics: Structures of intermediates and reaction mechanism from time-resolved x-ray data
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4J4Z
 
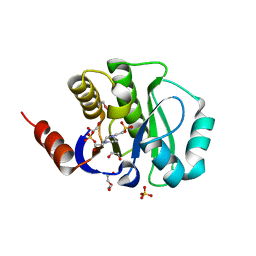 | | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Designed serine hydrolase variant OSH55.4_H1.2, ... | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301
To be Published
|
|
4JBC
 
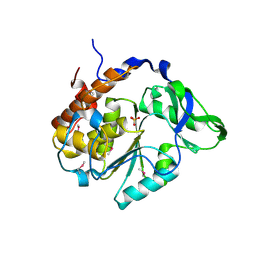 | | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318 | | Descriptor: | PHOSPHATE ION, designed serine hydrolase 3mmj_2 | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Baker, D, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318
To be Published
|
|
4K0C
 
 | | Crystal Structure of the computationally designed serine hydrolase. Northeast Structural Genomics Consortium (NESG) Target OR317 | | Descriptor: | designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR317
To be Published
|
|
4JGK
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275 | | Descriptor: | evolved variant of a designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275
To be Published
|
|
4N3P
 
 | | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Kogan, S, Maglaqui, M, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396
To be Published
|
|
3OEQ
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, with full length n-terminus | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3OER
 
 | | Crystal structure of trimeric frataxin from the yeast saccharomyces cerevisiae, complexed with cobalt | | Descriptor: | COBALT (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
4E3R
 
 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4GVV
 
 | | Crystal Structure of de novo design serine hydrolase OSH55.27, Northeast Structural Genomics Consortium (NESG) Target OR246 | | Descriptor: | De novo design serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR246
To be Published
|
|
4E3Q
 
 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4GVW
 
 | | Three-dimensional structure of the de novo designed serine hydrolase 2bfq_3, Northeast Structural Genomics Consortium (NESG) Target OR248 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, De novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR248
To be Published
|
|
