1M5C
 
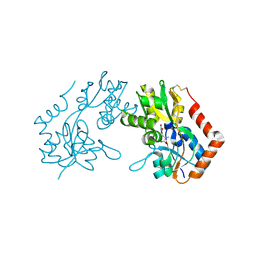 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
2AIX
 
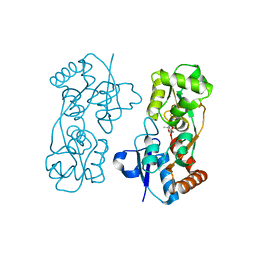 | |
7AFW
 
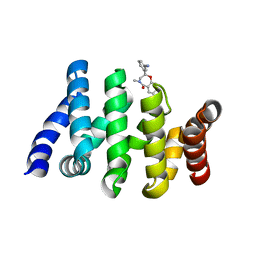 | | Beta-Catenin in complex with compound 6 | | Descriptor: | 3-[(2~{R})-4-methyl-5-oxidanylidene-2,3-dihydro-1,4-benzoxazepin-2-yl]benzenecarbonitrile, Catenin beta-1 | | Authors: | Boettcher, J, Kessler, D. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Getting a Grip on the Undrugged: Targeting beta-Catenin with Fragment-Based Methods.
Chemmedchem, 16, 2021
|
|
5MW2
 
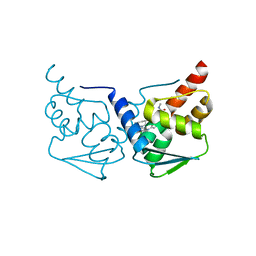 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
5MW6
 
 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
5MWD
 
 | | Crystal structure of the BCL6 BTB-domain with compound 2 | | Descriptor: | 5-[[5-chloranyl-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
2AL4
 
 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2,3,6A,7,8,9-HEXAHYDRO-11H-[1,4]DIOXINO[2,3-G]PYRROLO[2,1-B][1,3]BENZOXAZIN-11-ONE, Glutamate receptor 2, ... | | Authors: | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5D
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
9FWX
 
 | | Crystal structure of BRD4 BD1 with MEN1404BS. | | Descriptor: | 3-methyl-~{N}-(2-phenylethyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D. | | Deposit date: | 2024-07-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
9FXP
 
 | | Crystal structure of BRD4 BD1 with DI00626383. | | Descriptor: | 4-methoxy-1,2-benzoxazol-3-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Reinert, D. | | Deposit date: | 2024-07-02 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
7U8H
 
 | | Discovery of a KRAS G12V Inhibitor in vivo Tool Compound starting from an HSQC-NMR based Fragment Hit | | Descriptor: | 1H-benzimidazol-2-ylmethanethiol, 2-amino-4,5,6,7-tetrahydro-1-benzothiophene-3-carbonitrile, GTPase KRas, ... | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS G12C Inhibitor.
J.Med.Chem., 65, 2022
|
|
6RJ3
 
 | | Crystal structure of PHGDH in complex with compound 15 | | Descriptor: | 4-[(1~{R})-1-[(2-methyl-5-phenyl-pyrazol-3-yl)carbonylamino]ethyl]benzoic acid, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
2ANJ
 
 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
6RJ5
 
 | | Crystal structure of PHGDH in complex with compound 39 | | Descriptor: | 2-methyl-~{N}-[(1~{R})-1-[4-(methylsulfonylcarbamoyl)phenyl]ethyl]-5-phenyl-pyrazole-3-carboxamide, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RIH
 
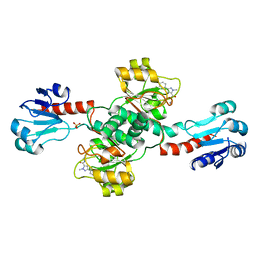 | | Crystal structure of PHGDH in complex with compound 9 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-cyclopropyl-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ2
 
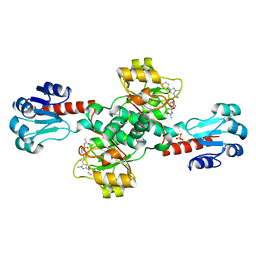 | | Crystal structure of PHGDH in complex with compound 40 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-[(1~{R})-1-[4-(ethanoylsulfamoyl)phenyl]ethyl]-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6S9B
 
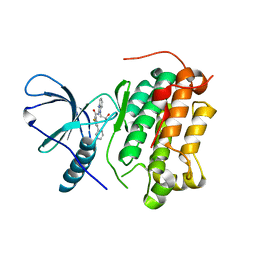 | | EGFR-KINASE IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 3-fluoranyl-~{N}-[1-(2-methyl-2-oxidanyl-propyl)benzimidazol-2-yl]-5-pyridin-3-yl-benzamide, Epidermal growth factor receptor | | Authors: | Bader, G. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Start Selective and Rigidify: The Discovery Path toward a Next Generation of EGFR Tyrosine Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6S9C
 
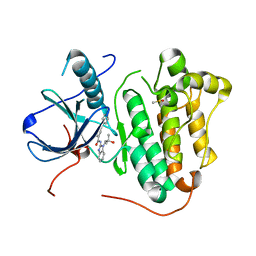 | | EGFR-KINASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | DIMETHYL SULFOXIDE, Epidermal growth factor receptor, ~{N}-[1-(2-methyl-2-oxidanyl-propyl)benzimidazol-2-yl]-2-phenyl-pyridine-4-carboxamide | | Authors: | Bader, G. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Start Selective and Rigidify: The Discovery Path toward a Next Generation of EGFR Tyrosine Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6S9D
 
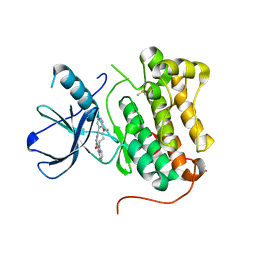 | | EGFR-KINASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 8-oxa-14,21,23,28-tetraazapentacyclo[23.3.1.02,7.014,22.015,20]nonacosa-1(28),2(7),3,5,15,17,19,21,25(29),26-decaen-24-one, DIMETHYL SULFOXIDE, Epidermal growth factor receptor | | Authors: | Bader, G. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Start Selective and Rigidify: The Discovery Path toward a Next Generation of EGFR Tyrosine Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
4ZWN
 
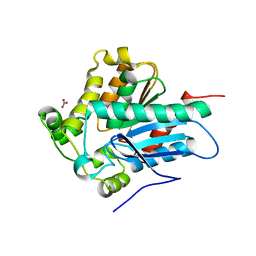 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
6RJ6
 
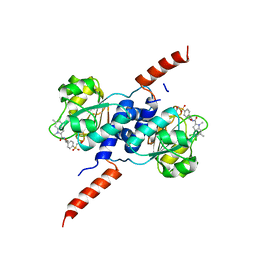 | | Crystal structure of PHGDH in complex with BI-4924 | | Descriptor: | 2-[4-[(1~{S})-1-[[4,5-bis(chloranyl)-1,6-dimethyl-indol-2-yl]carbonylamino]-2-oxidanyl-ethyl]phenyl]sulfonylethanoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
4ZXF
 
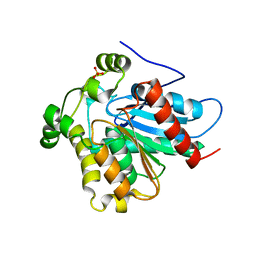 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | Descriptor: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
