5O4Y
 
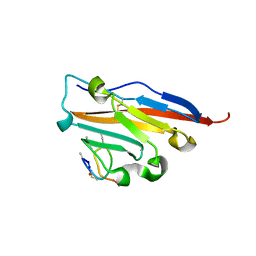 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | PHE-MAA-ASN-PRO-HIS-LEU-SER-TRP-SER-TRP-9KK-9KK-ARG-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | Authors: | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OAI
 
 | | Structure of MDM2 with low molecular weight inhibitor | | Descriptor: | 3-[(1~{R})-2-(~{tert}-butylamino)-1-[methanoyl-[[3,4,5-tris(fluoranyl)phenyl]methyl]amino]-2-oxidanylidene-ethyl]-6-chloranyl-1~{H}-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Neochoritis, C.G, Grudnik, P, Dubin, G, Domling, A, Holak, T.A. | | Deposit date: | 2017-06-22 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fluorinated indole-based MDM2 antagonist selectively inhibits the growth of p53wtosteosarcoma cells.
Febs J., 286, 2019
|
|
5N2D
 
 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[2,6-dimethoxy-4-[(2-methyl-3-phenyl-phenyl)methoxy]phenyl]methylamino]ethyl]ethanamide | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
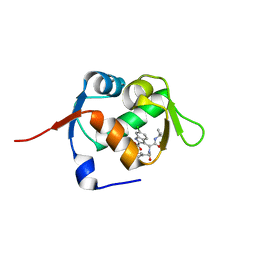
3PLU
 
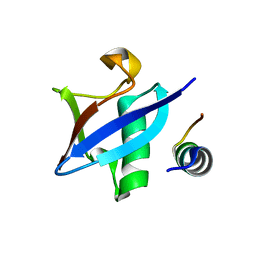 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDI) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
3PLV
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDII) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
5NIU
 
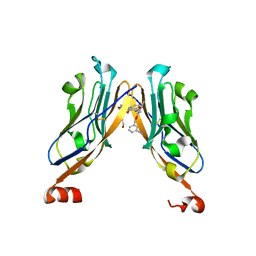 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{R})-2-[[2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-5-methyl-phenyl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells.
Oncotarget, 8, 2017
|
|
5N2F
 
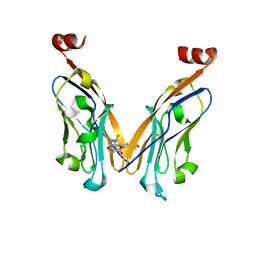 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
5N86
 
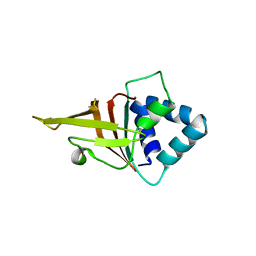 | | Crystal structure of FAS1 domain of hyaluronic acid receptor stabilin-2 | | Descriptor: | Stabilin-2 | | Authors: | Twarda-Clapa, A, Labuzek, B, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Crystal structure of the FAS1 domain of the hyaluronic acid receptor stabilin-2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5O45
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | PHE-MEA-9KK-SAR-ASP-VAL-MEA-TYR-SAR-TRP-TYR-LEU-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | Authors: | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-05-26 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3RMP
 
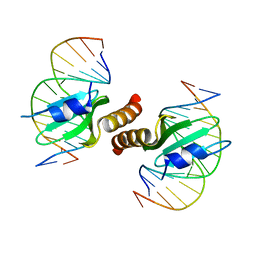 | | Structural basis for the recognition of attP substrates by P4-like integrases | | Descriptor: | 5'-D(*TP*AP*AP*TP*GP*AP*CP*CP*AP*CP*CP*AP*AP*TP*A)-3', 5'-D(*TP*AP*TP*TP*GP*GP*TP*GP*GP*TP*CP*AP*TP*TP*A)-3', CP4-like integrase | | Authors: | Szwagierczak, A, Popowicz, G.M, Holak, T.A, Rakin, A, Antonenka, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the recognition of attP substrates by P4-like integrases
To be Published
|
|
3TU1
 
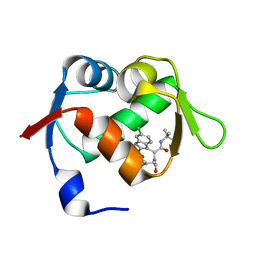 | | Exhaustive Fluorine Scanning towards Potent p53-MDM2 Antagonist | | Descriptor: | 3-[(1S)-2-(tert-butylamino)-1-{N-[(3,4-difluorophenyl)methyl]formamido}-2-oxoethyl]-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Wolf, S, Huang, Y, Koes, D, Popowicz, G.M, Camacho, C.J, Holak, T.A, Doemling, A. | | Deposit date: | 2011-09-15 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Exhaustive Fluorine Scanning toward Potent p53-Mdm2 Antagonists.
Chemmedchem, 7, 2012
|
|
3TC5
 
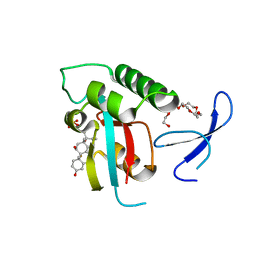 | | Selective targeting of disease-relevant protein binding domains by O-phosphorylated natural product derivatives | | Descriptor: | (11alpha,16alpha)-9-fluoro-11,17-dihydroxy-16-methyl-3,20-dioxopregna-1,4-dien-21-yl dihydrogen phosphate, HEXAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Graeber, M, Janczyk, W, Sperl, B, Elumalai, N, Kozany, C, Hausch, F, Holak, T.A, Berg, T. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective targeting of disease-relevant protein binding domains by o-phosphorylated natural product derivatives.
Acs Chem.Biol., 6, 2011
|
|
3TJ2
 
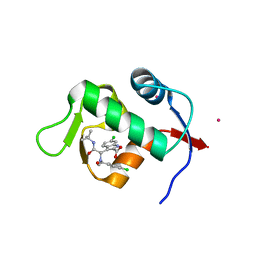 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[(4-chlorobenzyl)(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, POTASSIUM ION | | Authors: | Wolf, S, Huang, Y, Popowicz, G.M, Goda, S, Holak, T.A, Doemling, A. | | Deposit date: | 2011-08-23 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ugi Multicomponent Reaction Derived p53-Mdm2 Antagonists
To be published
|
|
3V3B
 
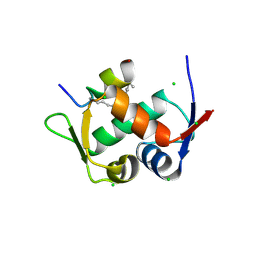 | | Structure of the Stapled p53 Peptide Bound to Mdm2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, SAH-p53-8 stapled-peptide | | Authors: | Baek, S, Kutchukian, P.S, Verdine, G.L, Huber, R, Holak, T.A, Ki Won, L, Popowicz, G.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the stapled p53 peptide bound to Mdm2.
J.Am.Chem.Soc., 134, 2012
|
|
3V3V
 
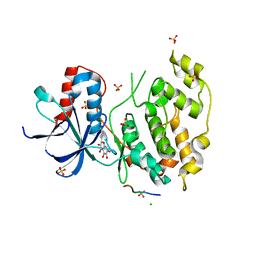 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
6R3K
 
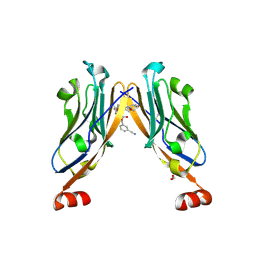 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[5-chloranyl-2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6RPG
 
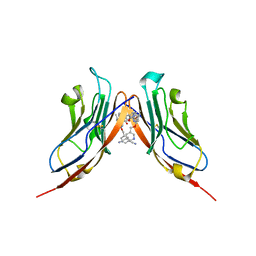 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | Authors: | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6SRU
 
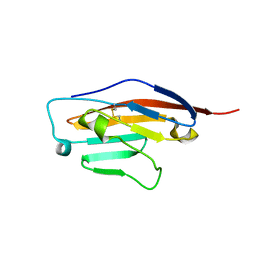 | | Structure of Ig-like V-type domian of mouse Programmed cell death 1 ligand 1 (PD-L1) | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Sala, D, Grudnik, P, Holak, T.A. | | Deposit date: | 2019-09-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Human and mouse PD-L1: similar molecular structure, but different druggability profiles.
Iscience, 24, 2021
|
|
2DSQ
 
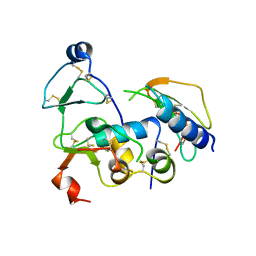 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 1, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GRC
 
 | | 1.5 A structure of bromodomain from human BRG1 protein, a central ATPase of SWI/SNF remodeling complex | | Descriptor: | Probable global transcription activator SNF2L4 | | Authors: | Singh, M, Popowicz, G.M, Krajewski, M, Holak, T.A. | | Deposit date: | 2006-04-24 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural ramification for acetyl-lysine recognition by the bromodomain of human BRG1 protein, a central ATPase of the SWI/SNF remodeling complex.
Chembiochem, 8, 2007
|
|
2DSP
 
 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DSR
 
 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1YZ5
 
 | |
2AS9
 
 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | Descriptor: | ZINC ION, serine protease | | Authors: | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | Deposit date: | 2005-08-23 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
1ZLI
 
 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with human carboxypeptidase B | | Descriptor: | Carboxypeptidase B, ZINC ION, carboxypeptidase inhibitor | | Authors: | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | Deposit date: | 2005-05-06 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
